Philip Dalsbecker, Siiri Suominen, Muhammad Asim Faridi, Reza Mahdavi, Julia Johansson, Charlotte Hamngren Blomqvist, Mattias Goksör, Katriina Aalto-Setälä, Leena E. Viiri and Caroline B. Adiels
Lab on a Chip 25, 4328 – 4344 (2025)
doi: 10.1039/D4LC00509K
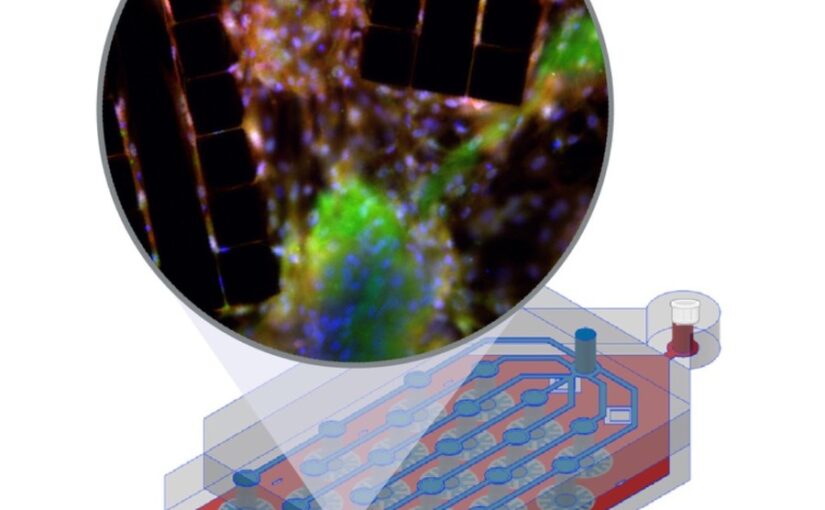
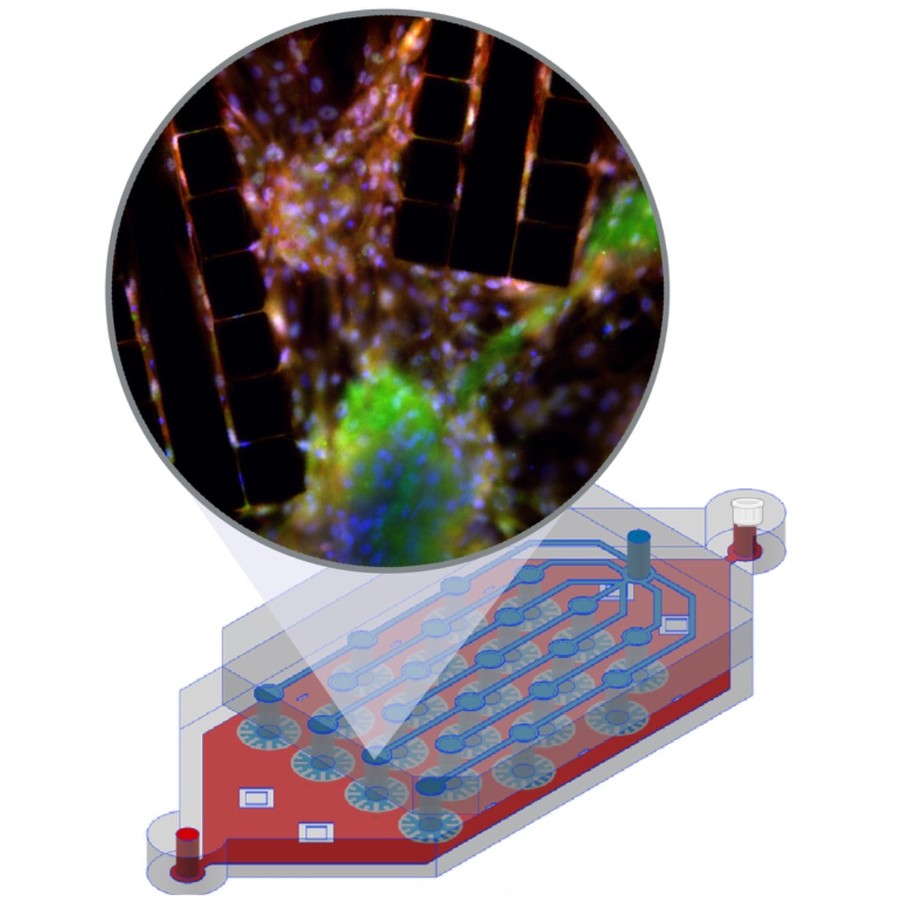
In vitro cell culture models play a crucial role in preclinical drug discovery. To achieve optimal culturing environments and establish physiologically relevant organ-specific conditions, it is imperative to replicate in vivo scenarios when working with primary or induced pluripotent cell types. However, current approaches to recreating in vivo conditions and generating relevant 3D cell cultures still fall short. In this study, we validate a liver-lobule-chip (LLoC) containing 21 artificial liver lobules, each representing the smallest functional unit of the human liver. The LLoC facilitates diffusion-based perfusion via sinusoid-mimetic structures, providing physiologically relevant shear stress exposure and radial nutrient concentration gradients within each lobule. We demonstrate the feasibility of long term cultures (up to 14 days) of viable and functional HepG2 cells in a 3D discoid tissue structure, serving as initial proof of concept. Thereafter, we successfully differentiate sensitive, human induced pluripotent stem cell (iPSC)-derived cells into hepatocyte-like cells over a period of 20 days on-chip, exhibiting advancements in maturity compared to traditional 2D cultures. Further, hepatocyte-like cells cultured in the LLoC exhibit zonated protein expression profiles, indicating the presence of metabolic gradients characteristic of liver lobules. Our results highlight the suitability of the LLoC for long-term discoid tissue cultures, specifically for iPSCs, and their differentiation in a perfused environment. We envision the LLoC as a starting point for more advanced in vitro models, allowing for the combination of multiple liver cell types to create a comprehensive liver model for disease-onchip studies. Ultimately, when combined with stem cell technology, the LLoC offers a promising and robust on-chip liver model that serves as a viable alternative to primary hepatocyte cultures—ideally suited for preclinical drug screening and personalized medicine applications.