Erik Olsén, Berenice García Rodríguez, Fredrik Skärberg, Petteri Parkkila, Giovanni Volpe, Fredrik Höök, and Daniel Sundås Midtvedt
Nano Letters (2024)
arXiv: 2309.07572
doi: 10.1021/acs.nanolett.3c03539
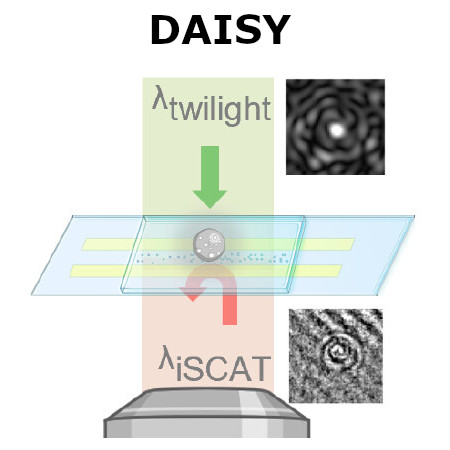
Traditional single-nanoparticle sizing using optical microscopy techniques assesses size via the diffusion constant, which requires suspended particles to be in a medium of known viscosity. However, these assumptions are typically not fulfilled in complex natural sample environments. Here, we introduce dual-angle interferometric scattering microscopy (DAISY), enabling optical quantification of both size and polarizability of individual nanoparticles (radius <170 nm) without requiring a priori information regarding the surrounding media or super-resolution imaging. DAISY achieves this by combining the information contained in concurrently measured forward and backward scattering images through twilight off-axis holography and interferometric scattering (iSCAT). Going beyond particle size and polarizability, single-particle morphology can be deduced from the fact that the hydrodynamic radius relates to the outer particle radius, while the scattering-based size estimate depends on the internal mass distribution of the particles. We demonstrate this by differentiating biomolecular fractal aggregates from spherical particles in fetal bovine serum at the single-particle level.