Harshith Bachimanchi, Matthew I. M. Pinder, Chloé Robert, Pierre De Wit, Jonathan Havenhand, Alexandra Kinnby, Daniel Midtvedt, Erik Selander, Giovanni Volpe
Limnology and Oceanography Letters (2024)
doi: 10.1002/lol2.10392
arXiv: 2309.08500
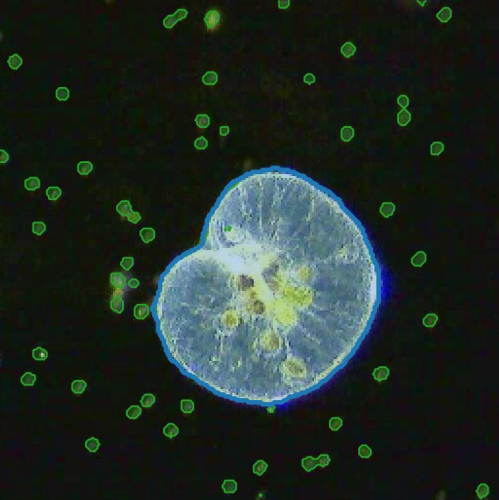
The implementation of deep learning algorithms has brought new perspectives to plankton ecology. Emerging as an alternative approach to established methods, deep learning offers objective schemes to investigate plankton organisms in diverse environments. We provide an overview of deep-learning-based methods including detection and classification of phytoplankton and zooplankton images, foraging and swimming behavior analysis, and finally ecological modeling. Deep learning has the potential to speed up the analysis and reduce the human experimental bias, thus enabling data acquisition at relevant temporal and spatial scales with improved reproducibility. We also discuss shortcomings and show how deep learning architectures have evolved to mitigate imprecise readouts. Finally, we suggest opportunities where deep learning is particularly likely to catalyze plankton research. The examples are accompanied by detailed tutorials and code samples that allow readers to apply the methods described in this review to their own data.