Label-free characterization of biological matter across scales
Daniel Midtvedt, Erik Olsén, Benjamin Midtvedt, Elin K. Esbjörner, Fredrik Skärberg, Berenice Garcia, Caroline B. Adiels, Fredrik Höök, Giovanni Volpe
SPIE-ETAI
Date: 24 August 2022
Time: 09:10 (PDT)
Tag: Daniel Midtvedt
Presentation by D. Midtvedt at SPIE-ETAI, San Diego, 23 August 2022
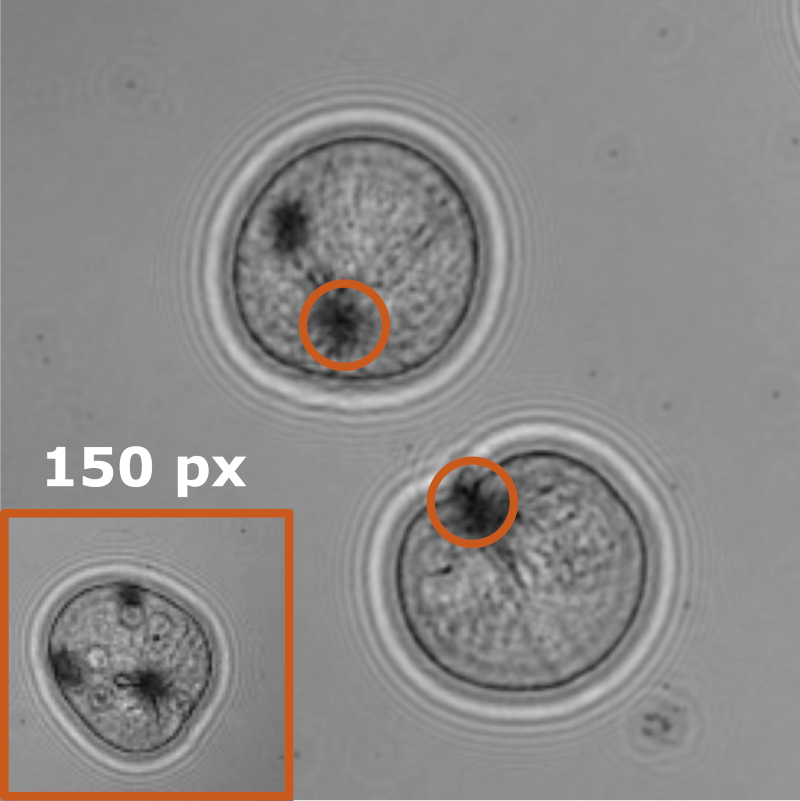
Benjamin Midtvedt, Jesus Pineda, Fredrik Skärberg, Erik Olsén, Harshith Bachimanchi, Emelie Wesén, Elin Esbjörner, Erik Selander, Fredrik Höök, Daniel Midtvedt, Giovanni Volpe
Submitted to SPIE-ETAI
Date: 23 August 2022
Time: 2:20 PM (PDT)
Object detection is a fundamental task in digital microscopy. Recently, machine-learning approaches have made great strides in overcoming the limitations of more classical approaches. The training of state-of-the-art machine-learning methods almost universally relies on either vast amounts of labeled experimental data or the ability to numerically simulate realistic datasets. However, the data produced by experiments are often challenging to label and cannot be easily reproduced numerically. Here, we propose a novel deep-learning method, named LodeSTAR (Low-shot deep Symmetric Tracking And Regression), that learns to detect small, spatially confined, and largely homogeneous objects that have sufficient contrast to the background with sub-pixel accuracy from a single unlabeled experimental image. This is made possible by exploiting the inherent roto-translational symmetries of the data. We demonstrate that LodeSTAR outperforms traditional methods in terms of accuracy. Furthermore, we analyze challenging experimental data containing densely packed cells or noisy backgrounds. We also exploit additional symmetries to extend the measurable particle properties to the particle’s vertical position by propagating the signal in Fourier space and its polarizability by scaling the signal strength. Thanks to the ability to train deep-learning models with a single unlabeled image, LodeSTAR can accelerate the development of high-quality microscopic analysis pipelines for engineering, biology, and medicine.
Soft Matter Lab members present at SPIE Optics+Photonics conference in San Diego, 21-25 August 2022
The Soft Matter Lab participates to the SPIE Optics+Photonics conference in San Diego, CA, USA, 21-25 August 2022, with the presentations listed below.
- Martin Selin: Scalable construction of quantum dot arrays using optical tweezers and deep learning (@SPIE)
22 August 2022 • 11:05 AM – 11:25 AM PDT | Conv. Ctr. Room 5A
- Fredrik Skärberg: Holographic characterisation of biological nanoparticles using deep learning (@SPIE)
22 August 2022 • 5:30 PM – 7:30 PM PDT | Conv. Ctr. Exhibit Hall B1
- Zofia Korczak: Dynamic virtual live/apoptotic cell assay using deep learning (@SPIE)
22 August 2022 • 5:30 PM – 7:30 PM PDT | Conv. Ctr. Exhibit Hall B1
- Henrik Klein Moberg: Seeing the invisible: deep learning optical microscopy for label-free biomolecule screening in the sub-10 kDa regime (@SPIE)
23 August 2022 • 9:15 AM – 9:35 AM PDT | Conv. Ctr. Room 5A
- Jesus Pineda: Revealing the spatiotemporal fingerprint of microscopic motion using geometric deep learning (@SPIE)
23 August 2022 • 11:05 AM – 11:25 AM PDT | Conv. Ctr. Room 5A
- Agnese Callegari: Simulating intracavity optical trapping with machine learning (@SPIE)
23 August 2022 • 1:40 PM – 2:00 PM PDT | Conv. Ctr. Room 5A
- Benjamin Midtvedt: Single-shot self-supervised particle tracking (@SPIE)
23 August 2022 • 2:20 PM – 2:40 PM PDT | Conv. Ctr. Room 5A
- Yu-Wei Chang: Neural network training with highly incomplete medical datasets (@SPIE)
24 August 2022 • 8:00 AM – 8:20 AM PDT | Conv. Ctr. Room 5A
- Daniel Midtvedt: Label-free characterization of biological matter across scales (@SPIE)
24 August 2022 • 9:10 AM – 9:40 AM PDT | Conv. Ctr. Room 5A
- Harshith Bachimanchi: Quantitative microplankton tracking by holographic microscopy and deep learning (@SPIE)
24 August 2022 • 11:45 AM – 12:05 PM PDT | Conv. Ctr. Room 5A
- Pablo Emiliano Gomez Ruiz: BRAPH 2.0: brain connectivity software with multilayer network analyses and machine learning (@SPIE)
24 August 2022 • 2:05 PM – 2:25 PM PDT | Conv. Ctr. Room 5A
- Yu-Wei Chang: Deep-learning analysis in tau PET for Alzheimer’s continuum (@SPIE)
24 August 2022 • 4:40 PM – 5:00 PM PDT | Conv. Ctr. Room 5A
Giovanni Volpe is also co-author of the presentations:
- Antonio Ciarlo: Periodic feedback effect in counterpropagating intracavity optical tweezers (@SPIE)
24 August 2022 • 2:00 PM – 2:20 PM PDT | Conv. Ctr. Room 1B
- Anna Canal Garcia: Multilayer brain connectivity analysis in Alzheimer’s disease using functional MRI data (@SPIE)
24 August 2022 • 2:25 PM – 2:45 PM PDT | Conv. Ctr. Room 5A
- Mite Mijalkov: A novel method for quantifying men and women-like features in brain structure and function (@SPIE)
24 August 2022 • 3:05 PM – 3:25 PM PDT | Conv. Ctr. Room 5A
- Carlo Manzo: An anomalous competition: assessment of methods for anomalous diffusion through a community effort (@SPIE)
25 August 2022 • 9:00 AM – 9:30 AM PDT | Conv. Ctr. Room 5A
- Stefano Bo: Characterization of anomalous diffusion with neural networks (@SPIE)
25 August 2022 • 10:00 AM – 10:30 AM PDT | Conv. Ctr. Room 5A
Deep learning in light–matter interactions published in Nanophotonics
Daniel Midtvedt, Vasilii Mylnikov, Alexander Stilgoe, Mikael Käll, Halina Rubinsztein-Dunlop and Giovanni Volpe
Nanophotonics, 11(14), 3189-3214 (2022)
doi: 10.1515/nanoph-2022-0197
The deep-learning revolution is providing enticing new opportunities to manipulate and harness light at all scales. By building models of light–matter interactions from large experimental or simulated datasets, deep learning has already improved the design of nanophotonic devices and the acquisition and analysis of experimental data, even in situations where the underlying theory is not sufficiently established or too complex to be of practical use. Beyond these early success stories, deep learning also poses several challenges. Most importantly, deep learning works as a black box, making it difficult to understand and interpret its results and reliability, especially when training on incomplete datasets or dealing with data generated by adversarial approaches. Here, after an overview of how deep learning is currently employed in photonics, we discuss the emerging opportunities and challenges, shining light on how deep learning advances photonics.
Label-free nanofluidic scattering microscopy of size and mass of single diffusing molecules and nanoparticles published in Nature Methods
Barbora Špačková, Henrik Klein Moberg, Joachim Fritzsche, Johan Tenghamn, Gustaf Sjösten, Hana Šípová-Jungová, David Albinsson, Quentin Lubart, Daniel van Leeuwen, Fredrik Westerlund, Daniel Midtvedt, Elin K. Esbjörner, Mikael Käll, Giovanni Volpe & Christoph Langhammer
Nature Methods 19, 751–758 (2022)
doi: 10.1038/s41592-022-01491-6
Label-free characterization of single biomolecules aims to complement fluorescence microscopy in situations where labeling compromises data interpretation, is technically challenging or even impossible. However, existing methods require the investigated species to bind to a surface to be visible, thereby leaving a large fraction of analytes undetected. Here, we present nanofluidic scattering microscopy (NSM), which overcomes these limitations by enabling label-free, real-time imaging of single biomolecules diffusing inside a nanofluidic channel. NSM facilitates accurate determination of molecular weight from the measured optical contrast and of the hydrodynamic radius from the measured diffusivity, from which information about the conformational state can be inferred. Furthermore, we demonstrate its applicability to the analysis of a complex biofluid, using conditioned cell culture medium containing extracellular vesicles as an example. We foresee the application of NSM to monitor conformational changes, aggregation and interactions of single biomolecules, and to analyze single-cell secretomes.
Seminar by D. Midtvedt at Freie Universität Berlin, 29 October 2021

Daniel Midtvedt
(online at) Freie Universität Berlin, Germany
29 October 2021
Video microscopy has a long history of providing insight and breakthroughs for a broad range of disciplines, from physics to biology. Image analysis to extract quantitative information from video microscopy data has traditionally relied on algorithmic approaches, which are often difficult to implement, time-consuming, and computationally expensive. Recently, alternative data-driven approaches using deep learning have greatly improved quantitative digital microscopy, potentially offering automatized, accurate, and fast image analysis.
However, the combination of deep learning and video microscopy remains underutilized primarily due to the steep learning curve involved in developing custom deep-learning solutions. To overcome this issue, we recently introduced a software, DeepTrack 2.0, to design, train, and validate deep-learning solutions for digital microscopy.
In this talk, I will show how this software can be used in a broad range of applications, from particle localization, tracking, and characterization, to cell counting and classification. Thanks to its user-friendly graphical interface, DeepTrack 2.0 can be easily customized for user-specific applications, and thanks to its open-source, object-oriented programing, it can be easily expanded to add features and functionalities, potentially introducing deep-learning-enhanced video microscopy to a far wider audience.
Press release on Extracting quantitative biological information from bright-field cell images using deep learning
The article Extracting quantitative biological information from bright-field cell images using deep learning has been featured in a press release of the University of Gothenburg.
The study, recently published in Biophysics Reviews, shows how artificial intelligence can be used to develop faster, cheaper and more reliable information about cells, while also eliminating the disadvantages from using chemicals in the process.
Here the links to the press releases on Cision:
Swedish: Effektivare studier av celler med ny AI-metod
English: More effective cell studies using new AI method
Here the links to the press releases in the News of the University of Gothenburg:
Swedish: Effektivare studier av celler med ny AI-metod
English: More effective cell studies using new AI method
Extracting quantitative biological information from brightfield cell images using deep learning featured in AIP SciLight
The article Extracting quantitative biological information from brightfield cell images using deep learning
has been featured in: “Staining Cells Virtually Offers Alterative Approach to Chemical Dyes”, AIP SciLight (July 23, 2021).
Scilight showcases the most interesting research across the physical sciences published in AIP Publishing Journals.
Scilight is published weekly (52 issues per year) by AIP Publishing.
Extracting quantitative biological information from bright-field cell images using deep learning published in Biophysics Reviews

Saga Helgadottir, Benjamin Midtvedt, Jesús Pineda, Alan Sabirsh, Caroline B. Adiels, Stefano Romeo, Daniel Midtvedt, Giovanni Volpe
Biophysics Rev. 2, 031401 (2021)
arXiv: 2012.12986
doi: 10.1063/5.0044782
Quantitative analysis of cell structures is essential for biomedical and pharmaceutical research. The standard imaging approach relies on fluorescence microscopy, where cell structures of interest are labeled by chemical staining techniques. However, these techniques are often invasive and sometimes even toxic to the cells, in addition to being time-consuming, labor-intensive, and expensive. Here, we introduce an alternative deep-learning-powered approach based on the analysis of bright-field images by a conditional generative adversarial neural network (cGAN). We show that this approach can extract information from the bright-field images to generate virtually-stained images, which can be used in subsequent downstream quantitative analyses of cell structures. Specifically, we train a cGAN to virtually stain lipid droplets, cytoplasm, and nuclei using bright-field images of human stem-cell-derived fat cells (adipocytes), which are of particular interest for nanomedicine and vaccine development. Subsequently, we use these virtually-stained images to extract quantitative measures about these cell structures. Generating virtually-stained fluorescence images is less invasive, less expensive, and more reproducible than standard chemical staining; furthermore, it frees up the fluorescence microscopy channels for other analytical probes, thus increasing the amount of information that can be extracted from each cell.
Quantitative Digital Microscopy with Deep Learning published in Applied Physics Reviews
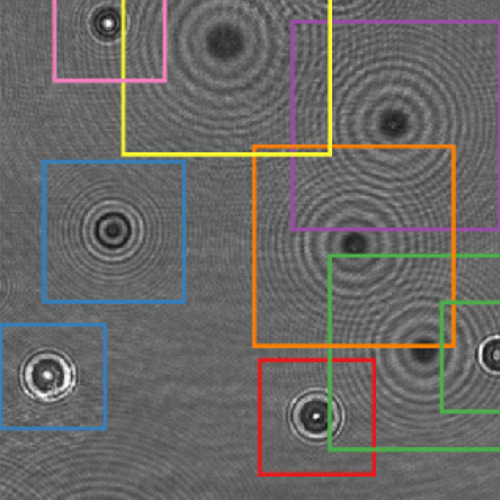
Quantitative Digital Microscopy with Deep Learning
Benjamin Midtvedt, Saga Helgadottir, Aykut Argun, Jesús Pineda, Daniel Midtvedt, Giovanni Volpe
Applied Physics Reviews 8, 011310 (2021)
doi: 10.1063/5.0034891
arXiv: 2010.08260
Video microscopy has a long history of providing insights and breakthroughs for a broad range of disciplines, from physics to biology. Image analysis to extract quantitative information from video microscopy data has traditionally relied on algorithmic approaches, which are often difficult to implement, time consuming, and computationally expensive. Recently, alternative data-driven approaches using deep learning have greatly improved quantitative digital microscopy, potentially offering automatized, accurate, and fast image analysis. However, the combination of deep learning and video microscopy remains underutilized primarily due to the steep learning curve involved in developing custom deep-learning solutions. To overcome this issue, we introduce a software, DeepTrack 2.0, to design, train and validate deep-learning solutions for digital microscopy. We use it to exemplify how deep learning can be employed for a broad range of applications, from particle localization, tracking and characterization to cell counting and classification. Thanks to its user-friendly graphical interface, DeepTrack 2.0 can be easily customized for user-specific applications, and, thanks to its open-source object-oriented programming, it can be easily expanded to add features and functionalities, potentially introducing deep-learning-enhanced video microscopy to a far wider audience.