Harshith Bachimanchi Benjamin Midtvedt, Daniel Midtvedt, Erik Selander, and Giovanni Volpe
Presentation at SPIE-ETAI 2022
San Diego, USA
24 August 2022, 11:45 PDT
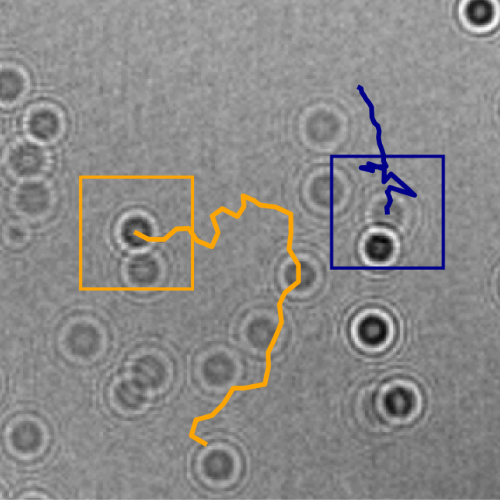
The marine microbial food web plays a central role in the global carbon cycle. Our mechanistic understanding of the ocean, however, is biased towards its larger constituents, while rates and biomass fluxes in the microbial food web are mainly inferred from indirect measurements and ensemble averages.Yet, resolution at the level of the individual microplankton is required to advance our understanding of the oceanic food web. Here, we demonstrate that, by combining holographic microscopy with deep learning, we can follow microplanktons through generations, continuously measuring their three dimensional position and dry mass. The deep learning algorithms circumvent the computationally intensive processing of holographic data and allow inline measurements over extended time periods. This permits us to reliably estimate growth rates, both in terms of dry mass increase and cell divisions, as well as to measure trophic interactions between species such as predation events. The individual resolution provides information about selectivity, individual feeding rates and handling times for individual microplanktons. This method is particularly useful to explore the flux of carbon through microzooplankton, the most important and least known group of primary consumers in the global oceans. We exemplify this by detailed descriptions of microzooplankton feeding events, cell divisions, and long term monitoring of single cells from division to division.