
Yihuai is a master student in Data Science and AI at Chalmers University of Technology.
During his time at the Soft Matter lab, he will be working on developing reinforcement learning algorithm for robotic applications.

Yu-Wei Chang, Massimiliano Passaretti
NEMES 2025, 24-26 September, 2025
Date: 25 September 2025
Time: 12:45 – 14:00
Place: Clarion Hotel Draken
This workshop begins with a practical introduction to graph theory, then guides participants through BRAPH 2 to build connectomes, compute graph measures, and run group comparisons, followed by a hands-on deep-learning pipeline. It demonstrates a unified GUI/command-line workflow, a unique architecture of BRAPH 2, helping participants move smoothly from the GUI to scripts. This workshop also guides participants to reproduce multiplex and deep-learning results on their computers from the BRAPH 2 bioRxiv preprint.
Yu-Wei Chang
NEMES 2025, 24-26 September, 2025
Date: 25 September 2025
Time: 09:00 – 09:45
Place: Clarion Hotel Draken
This plenary presents a practical framework for analysing neuroimaging data with network science and deep learning. It moves from modality-specific preprocessing to graph construction (single-layer and multiplex), then covers core graph measures, group inference, and brain-surface visualization, highlighting recent work from Associate Professor Joana B. Pereira’s group (Department of Clinical Neuroscience, Karolinska Institutet). It also introduces deep-learning pipelines for neuroimaging data: reservoir-computing memory capacity analysis, GapNet for handling missing data, and a robust feature-attribution method combined with SNP (single nucleotide polymorphism) information. The plenary concludes with the BRAPH 2 framework, which supports these pipelines and extends to other ongoing projects (e.g., light-sheet microscopy, Raman spectroscopy).

Matilda is a master student in Engineering Physics at Chalmers University of Technology.
During her time at the Soft Matter Lab, she will be working on developing self-supervised deep learning methods for analyzing microscopy data.

Alex Lech, Mirja Granfors, Benjamin Midtvedt, Jesús Pineda, Harshith Bachimanchi, Carlo Manzo, Giovanni Volpe
BNMI 2025, 19-22 August 2025, Gothenburg, Sweden
Date: 20 August 2025
Time: 15:15-19:00
Place: Wallenberg Conference Centre
DeepTrack2 is a flexible and scalable Python library designed for simulating microscopy data to generate high-quality synthetic datasets for training deep learning models. It supports a wide range of imaging modalities, including brightfield, fluorescence, darkfield, and holography, allowing users to simulate realistic experimental conditions with ease. Its modular architecture enables users to customize experimental setups, simulate a variety of objects, and incorporate optical aberrations, realistic experimental noise, and other user-defined effects, making it suitable for various research applications. DeepTrack2 is designed to be an accessible tool for researchers in fields that utilize image analysis and deep learning, as it removes the need for labor-intensive manual annotation through simulations. This helps accelerate the development of AI-driven methods for experiments by providing large volumes of data that is often required by deep learning models. DeepTrack2 has already been used for a number of applications in cell tracking, classifications tasks, segmentations and holographic reconstruction. Its flexible and scalable nature enables researchers to simulate a wide array of experimental conditions and scenarios with full control of features and parameters.
DeepTrack2 is available on GitHub, with extensive documentation, tutorials, and an active community for support and collaboration at https://github.com/DeepTrackAI/DeepTrack2.
References:
Digital video microscopy enhanced by deep learning.
Saga Helgadottir, Aykut Argun & Giovanni Volpe.
Optica, volume 6, pages 506-513 (2019).
Quantitative Digital Microscopy with Deep Learning.
Benjamin Midtvedt, Saga Helgadottir, Aykut Argun, Jesús Pineda, Daniel Midtvedt & Giovanni Volpe.
Applied Physics Reviews, volume 8, article number 011310 (2021).
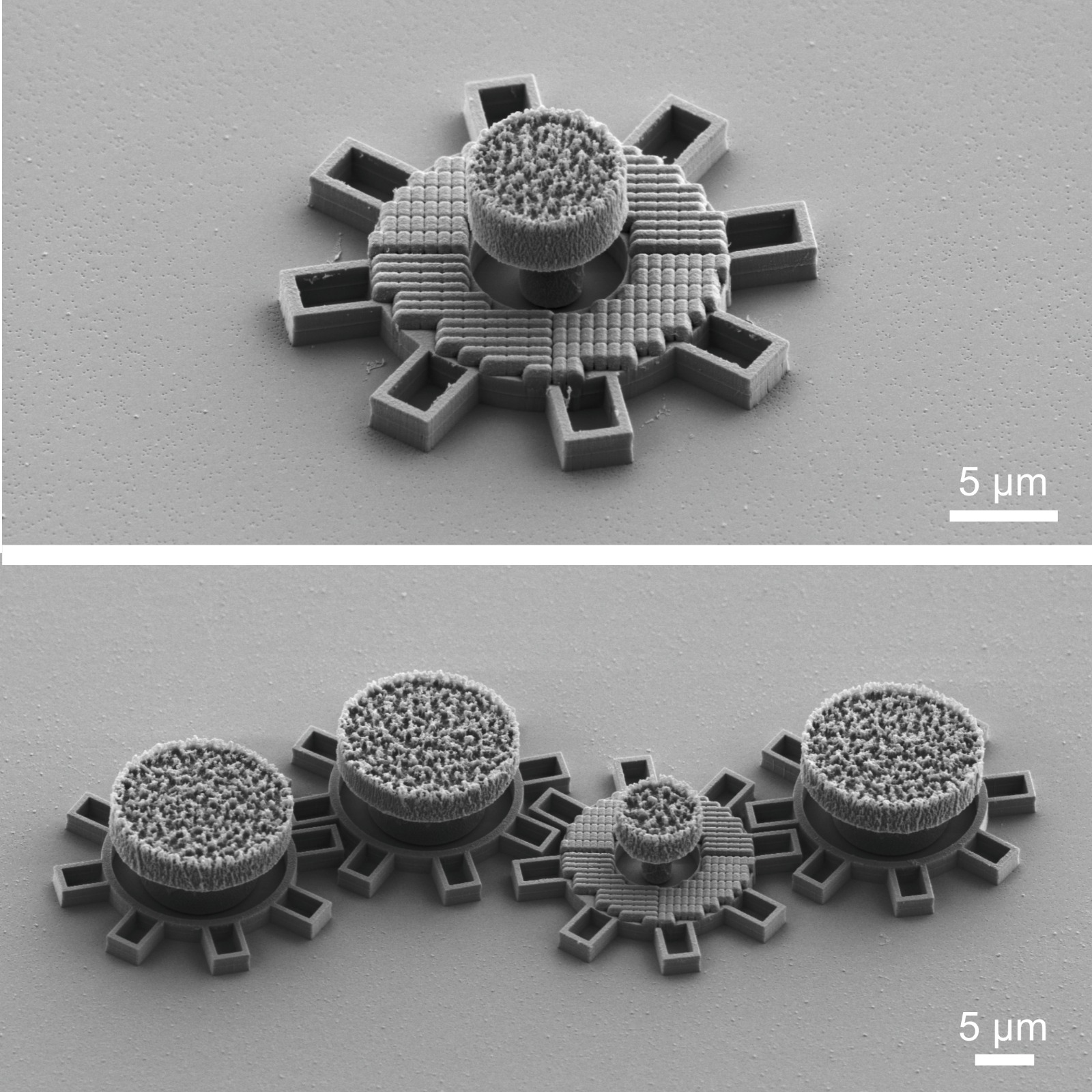
Microscopic Geared Metamachines
Gan Wang, Marcel Rey, Antonio Ciarlo, Mohanmmad Mahdi Shanei, Kunli Xiong, Giuseppe Pesce, Mikael Käll and Giovanni Volpe
Nature Communications 16, 7767 (2025)
doi: 10.1038/s41467-025-62869-6
arXiv: 2409.17284
The miniaturization of mechanical machines is critical for advancing nanotechnology and reducing device footprints. Traditional efforts to downsize gears and micromotors have faced limitations at around 0.1 mm for over thirty years due to the complexities of constructing drives and coupling systems at such scales. Here, we present an alternative approach utilizing optical metasurfaces to locally drive microscopic machines, which can then be fabricated using standard lithography techniques and seamlessly integrated on the chip, achieving sizes down to tens of micrometers with movements precise to the sub-micrometer scale. As a proof of principle, we demonstrate the construction of microscopic gear trains powered by a single driving gear with a metasurface activated by a plane light wave. Additionally, we develop a versatile pinion and rack micromachine capable of transducing rotational motion, performing periodic motion, and controlling microscopic mirrors for light deflection. Our on-chip fabrication process allows for straightforward parallelization and integration. Using light as a widely available and easily controllable energy source, these miniaturized metamachines offer precise control and movement, unlocking new possibilities for micro- and nanoscale systems.
After the article was published, it was reported by many media outlets, University of Gothenburg, New Scientist, Optics.org, Phys.org, ScienceDaily, Discover Magazine, among others.
Featured in:
GU: Light powered motor fits inside a strand of hair.
New Scientist: Microscopic gears powered by light could be used to make tiny machines and video on Youtube.
Optics.org: University of Gothenburg makes micron-scale light-powered gears
Phys.org: New light-powered gear fits inside a strand of hair
ScienceDaily: Scientists build micromotors smaller than a human hair
Discover Magazine: The Smallest Motors in History Can Fit Inside a Strand of Hair
@DrBenMiles: Scientists Create Micro Machines Powered by Light

Mirja Granfors, Alex Lech, Benjamin Midtvedt, Jesús Pineda, Harshith Bachimanchi, Carlo Manzo, and Giovanni Volpe
BNMI 2025, 19-22 August 2025, Gothenburg, Sweden
Date: 20 August 2025
Time: 15:00 – 15:15
Place: Wallenberg Conference Centre
DeepTrack2 is a flexible and scalable Python library designed to generate physics-based synthetic microscopy datasets for training deep learning models. It supports a wide range of imaging modalities, including brightfield, fluorescence, darkfield, and holography, enabling the creation of synthetic samples that accurately replicate real experimental conditions. Its modular architecture empowers users to customize optical systems, incorporate optical aberrations and noise, simulate diverse objects across various imaging scenarios, and apply image augmentations. DeepTrack2 is accompanied by a dedicated GitHub page, providing extensive documentation, examples, and an active community for support and collaboration: https://github.com/DeepTrackAI/DeepTrack2.
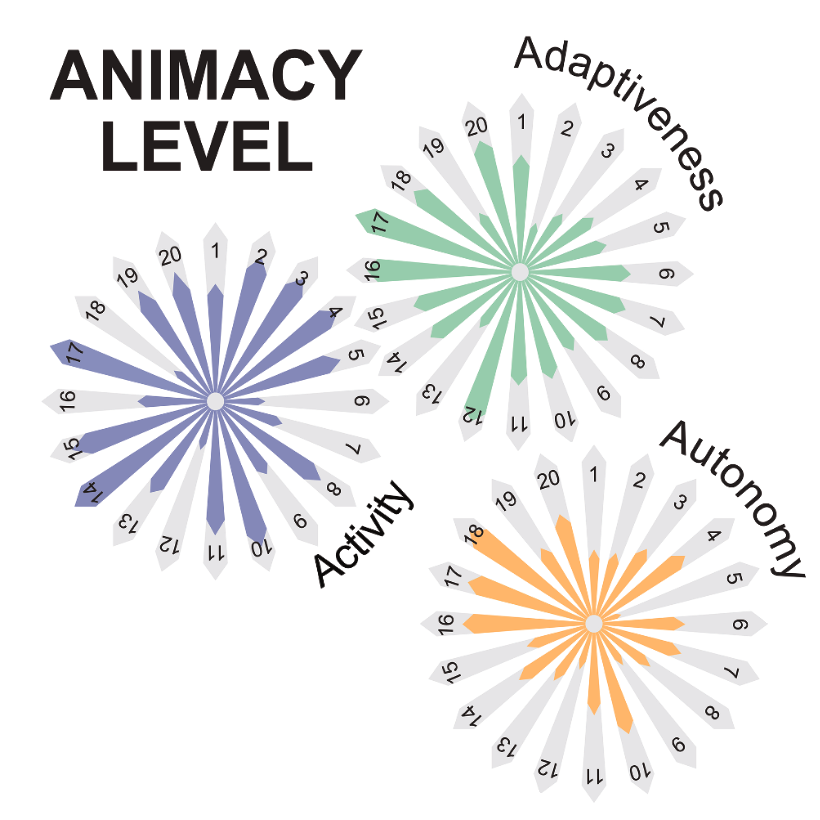
Giorgio Volpe, Nuno A M Araújo, Maria Guix, Mark Miodownik, Nicolas Martin, Laura Alvarez, Juliane Simmchen, Roberto Di Leonardo, Nicola Pellicciotta, Quentin Martinet, Jérémie Palacci, Wai Kit Ng, Dhruv Saxena, Riccardo Sapienza, Sara Nadine, João F Mano, Reza Mahdavi, Caroline Beck Adiels, Joe Forth, Christian Santangelo, Stefano Palagi, Ji Min Seok, Victoria A Webster-Wood, Shuhong Wang, Lining Yao, Amirreza Aghakhani, Thomas Barois, Hamid Kellay, Corentin Coulais, Martin van Hecke, Christopher J Pierce, Tianyu Wang, Baxi Chong, Daniel I Goldman, Andreagiovanni Reina, Vito Trianni, Giovanni Volpe, Richard Beckett, Sean P Nair, Rachel Armstrong
Journal of Physics: Condensed Matter 37, 333501 (2025)
arXiv: 2407.10623
doi: 10.1088/1361-648X/adebd3
Humanity has long sought inspiration from nature to innovate materials and devices. As science advances, nature-inspired materials are becoming part of our lives. Animate materials, characterized by their activity, adaptability, and autonomy, emulate properties of living systems. While only biological materials fully embody these principles, artificial versions are advancing rapidly, promising transformative impacts in the circular economy, health and climate resilience within a generation. This roadmap presents authoritative perspectives on animate materials across different disciplines and scales, highlighting their interdisciplinary nature and potential applications in diverse fields including nanotechnology, robotics and the built environment. It underscores the need for concerted efforts to address shared challenges such as complexity management, scalability, evolvability, interdisciplinary collaboration, and ethical and environmental considerations. The framework defined by classifying materials based on their level of animacy can guide this emerging field to encourage cooperation and responsible development. By unravelling the mysteries of living matter and leveraging its principles, we can design materials and systems that will transform our world in a more sustainable manner.

Jun Yi holds a bachelor’s degree in Chemistry from the University of Münster.
During his internship at the Soft Matter Lab, he will investigate the interactions of polymer-coated silica microparticles under various stimuli using optical tweezers.

Mirja Granfors received the Best Early Career Researcher Presentation Award at Emerging Topics in Artificial Intelligence (ETAI) 2025 held in San Diego, from 3 to 7 August 2025.
The award, which includes a certificate, a cash prize of $300, and a T-shirt, is presented by the organizers of the conference in collaboration with SPIE Optics + Photonics.
Mirja was awarded the prize for her presentation titled “DeepTrack2: physics-based microscopy simulations for deep learning”. Below is the full abstract of her presentation:
DeepTrack2 is a flexible and scalable Python library designed to generate physics-based synthetic microscopy datasets for training deep learning models. It supports a wide range of imaging modalities, including brightfield, fluorescence, darkfield, and holography, enabling the creation of synthetic samples that accurately replicate real experimental conditions. Its modular architecture empowers users to customize optical systems, incorporate optical aberrations and noise, simulate diverse objects across various imaging scenarios, and apply image augmentations. DeepTrack2 is accompanied by a dedicated GitHub page, providing extensive documentation, examples, and an active community for support and collaboration: https://github.com/DeepTrackAI/DeepTrack2.