Jesús Pineda, Sergi Masó-Orriols, Montse Masoliver, Joan Bertran, Mattias Goksör, Giovanni Volpe and Carlo Manzo
Nature Communications 16, 9693 (2025)
arXiv: 2412.00173
doi: 10.1038/s41467-025-65557-7
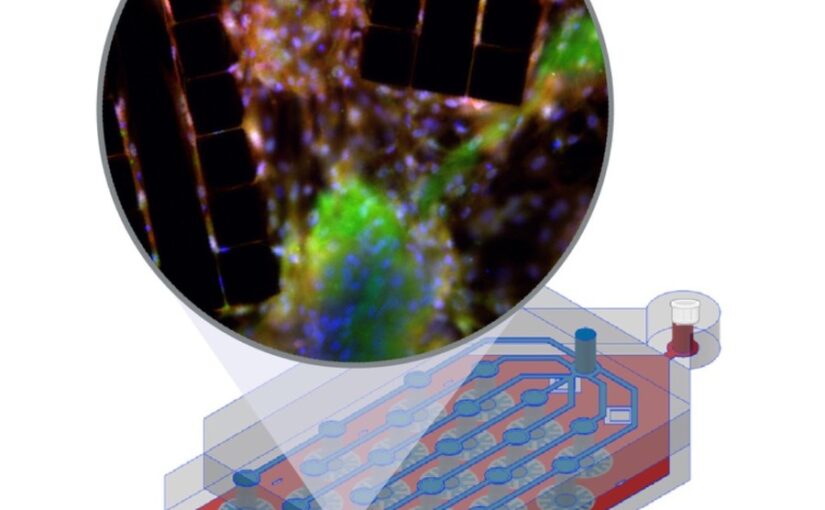
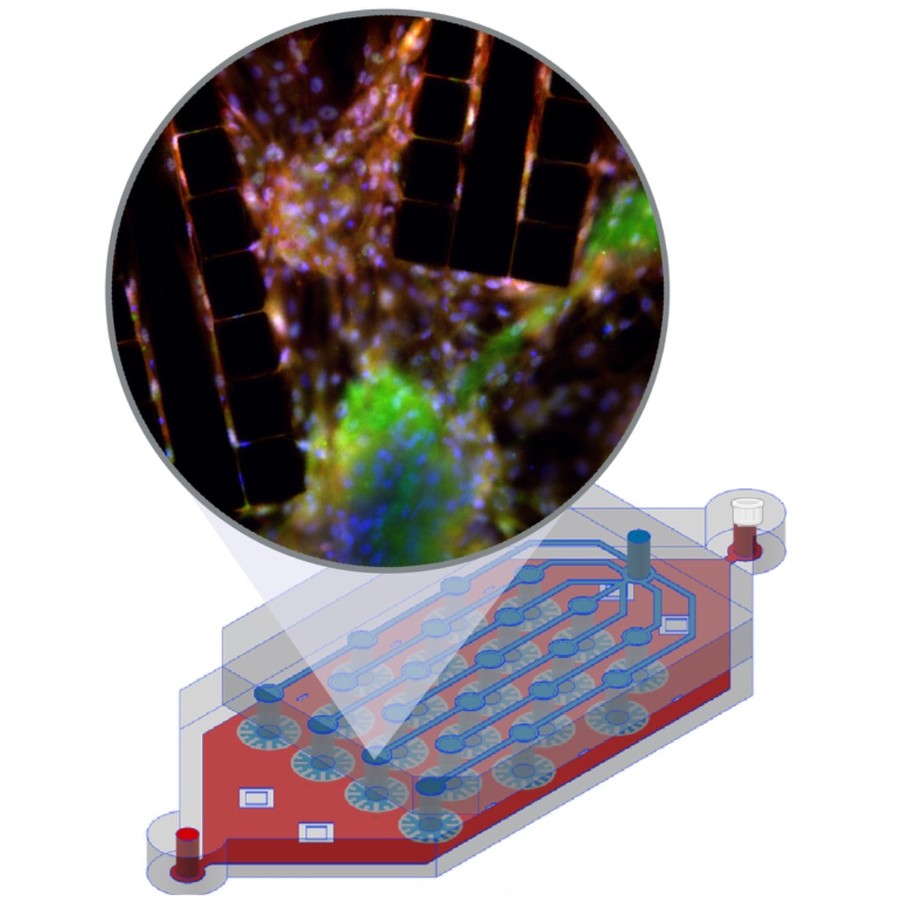
Single-molecule localization microscopy generates point clouds corresponding to fluorophore localizations. Spatial cluster identification and analysis of these point clouds are crucial for extracting insights about molecular organization. However, this task becomes challenging in the presence of localization noise, high point density, or complex biological structures. Here, we introduce MIRO (Multifunctional Integration through Relational Optimization), an algorithm that uses recurrent graph neural networks to transform the point clouds in order to improve clustering efficiency when applying conventional clustering techniques. We show that MIRO supports simultaneous processing of clusters of different shapes and at multiple scales, demonstrating improved performance across varied datasets. Our comprehensive evaluation demonstrates MIRO’s transformative potential for single-molecule localization applications, showcasing its capability to revolutionize cluster analysis and provide accurate, reliable details of molecular architecture. In addition, MIRO’s robust clustering capabilities hold promise for applications in various fields such as neuroscience, for the analysis of neural connectivity patterns, and environmental science, for studying spatial distributions of ecological data.