Giovanni Volpe
BIST Symposium on Microscopy, Nanoscopy and Imaging Sciences
Castedefells, 10 March 2023
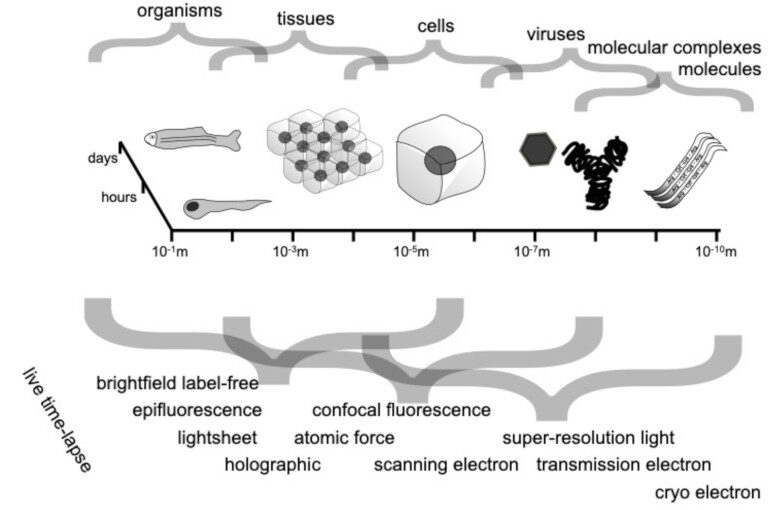
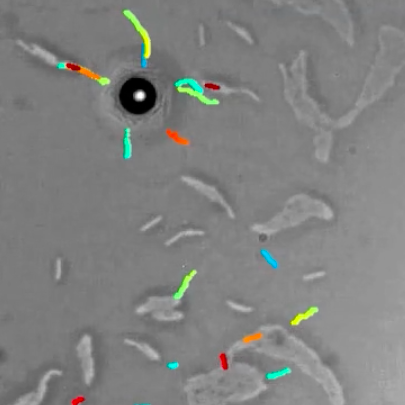
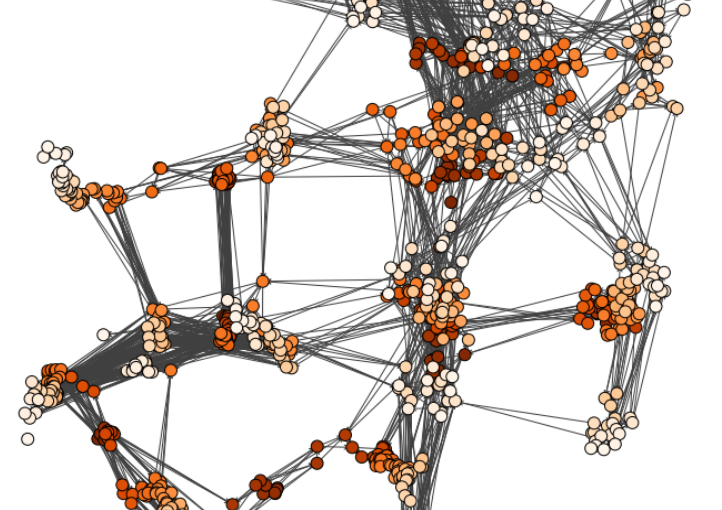
Video microscopy has a long history of providing insights and breakthroughs for a broad range of disciplines, from physics to biology. Image analysis to extract quantitative information from video microscopy data has traditionally relied on algorithmic approaches, which are often difficult to implement, time consuming, and computationally expensive. Recently, alternative data-driven approaches using deep learning have greatly improved quantitative digital microscopy, potentially offering automatized, accurate, and fast image analysis. However, the combination of deep learning and video microscopy remains underutilized primarily due to the steep learning curve involved in developing custom deep-learning solutions.
To overcome this issue, we have introduced a software, currently at version DeepTrack 2.1, to design, train and validate deep-learning solutions for digital microscopy. We use it to exemplify how deep learning can be employed for a broad range of applications, from particle localization, tracking and characterization to cell counting and classification. Thanks to its user-friendly graphical interface, DeepTrack 2.1 can be easily customized for user-specific applications, and, thanks to its open-source object-oriented programming, it can be easily expanded to add features and functionalities, potentially introducing deep-learning-enhanced video microscopy to a far wider audience.