Yu-Wei Chang, Laura Natali, Oveis Jamialahmadi, Stefano Romeo, Joana B. Pereira, Giovanni Volpe
Machine Learning: Science and Technology 3, 035001 (2022)
arXiV: 2107.00429
doi: 10.1088/2632-2153/ac7b69
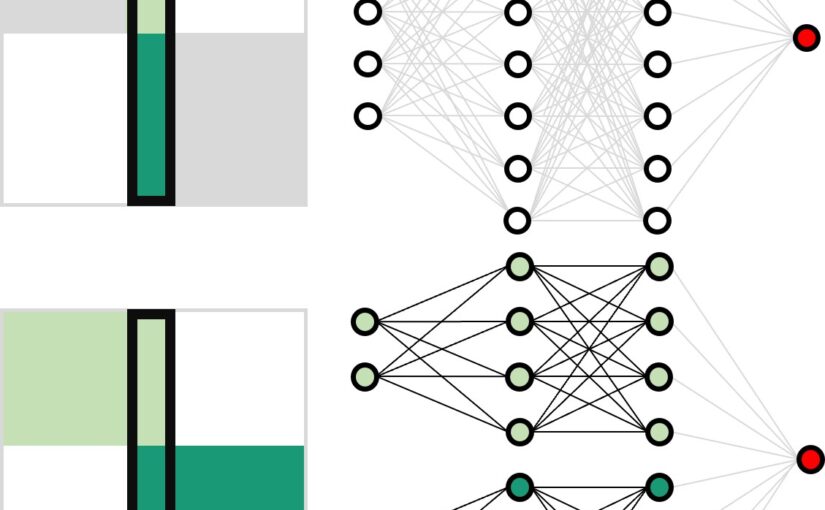
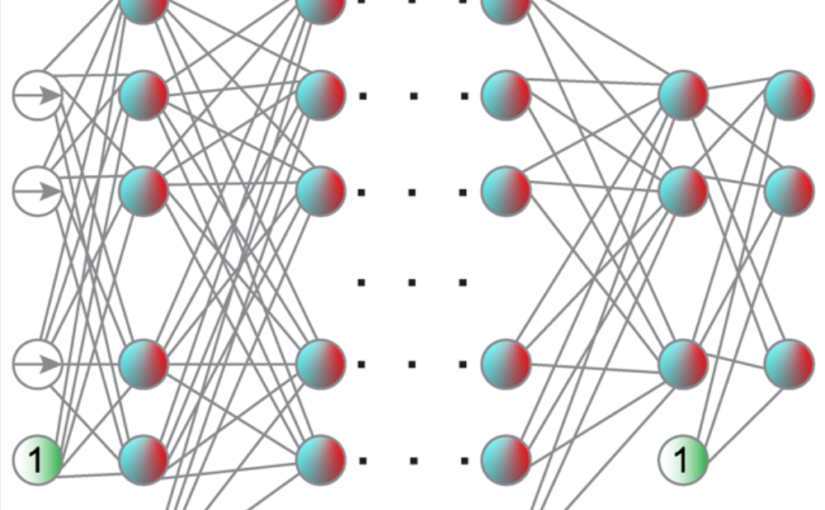
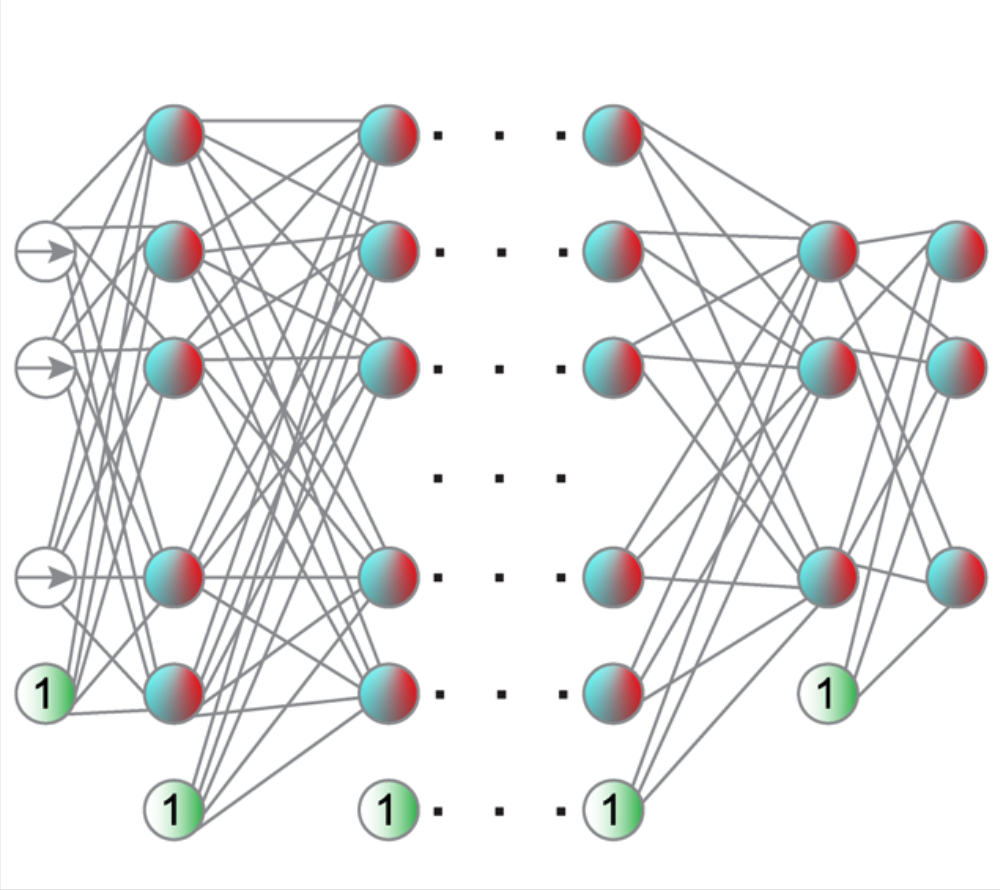
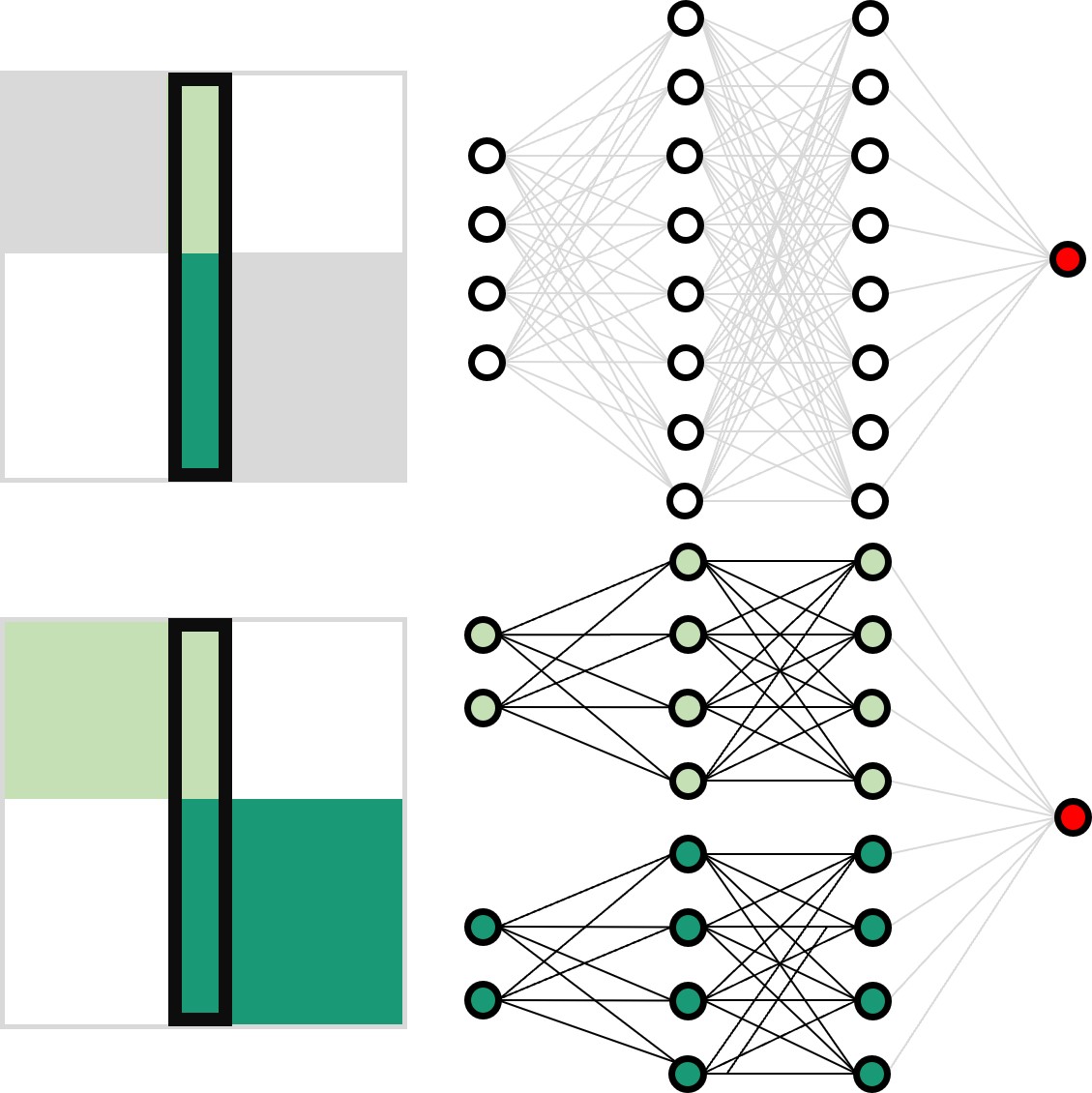
Neural network training and validation rely on the availability of large high-quality datasets. However, in many cases only incomplete datasets are available, particularly in health care applications, where each patient typically undergoes different clinical procedures or can drop out of a study. Since the data to train the neural networks need to be complete, most studies discard the incomplete datapoints, which reduces the size of the training data, or impute the missing features, which can lead to artefacts. Alas, both approaches are inadequate when a large portion of the data is missing. Here, we introduce GapNet, an alternative deep-learning training approach that can use highly incomplete datasets. First, the dataset is split into subsets of samples containing all values for a certain cluster of features. Then, these subsets are used to train individual neural networks. Finally, this ensemble of neural networks is combined into a single neural network whose training is fine-tuned using all complete datapoints. Using two highly incomplete real-world medical datasets, we show that GapNet improves the identification of patients with underlying Alzheimer’s disease pathology and of patients at risk of hospitalization due to Covid-19. By distilling the information available in incomplete datasets without having to reduce their size or to impute missing values, GapNet will permit to extract valuable information from a wide range of datasets, benefiting diverse fields from medicine to engineering.