Giovanni Volpe and Carlo Manzo
Computational Intelligence Group (CIG), Weekly Reading Session
Date: 3 July 2025
Time: 17:00
Place: Makerere University, Kampala, Uganda (Online)
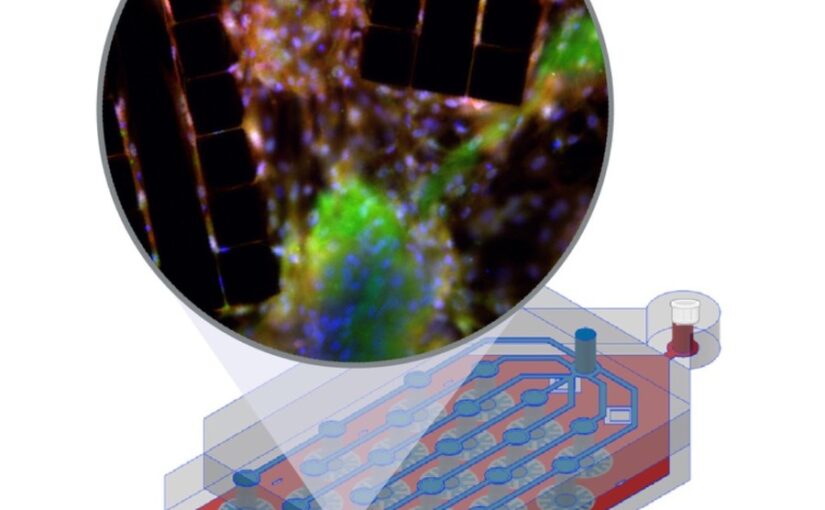
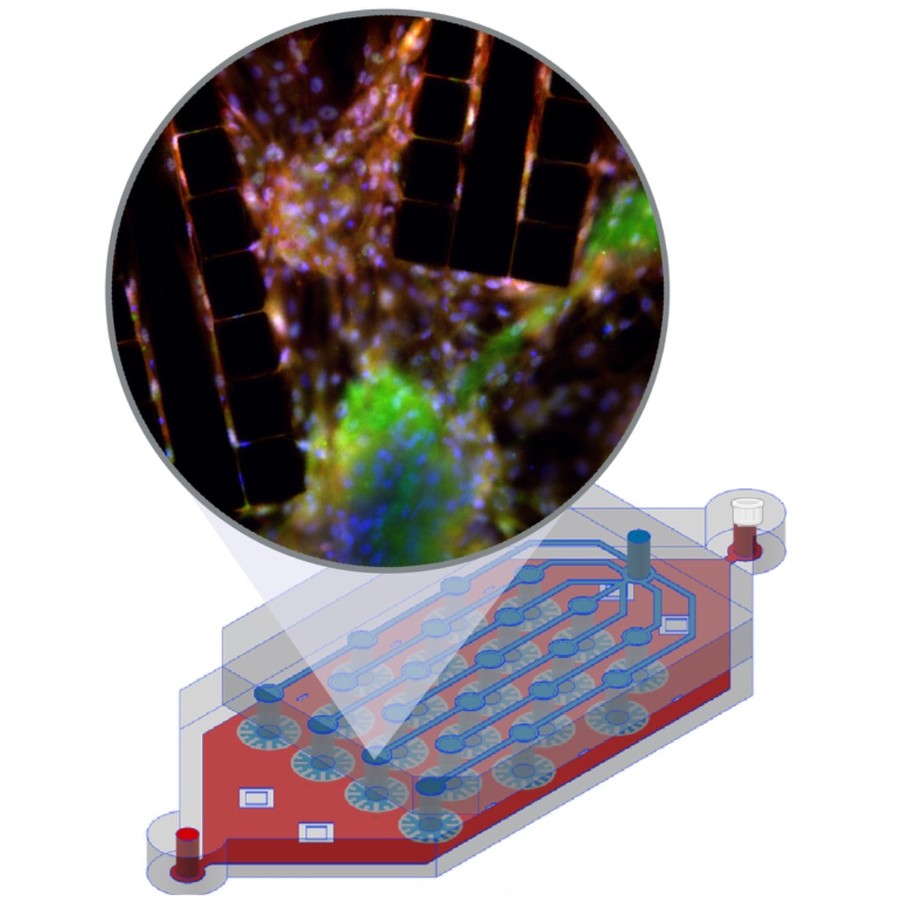
Graphs provide a powerful framework for modeling complex systems, but their structural variability makes analysis and classification challenging. To address this, we introduce GAUDI (Graph Autoencoder Uncovering Descriptive Information), a novel unsupervised geometric deep learning framework that captures both local details and global structure. GAUDI employs an innovative hourglass architecture with hierarchical pooling and upsampling layers, linked through skip connections to preserve essential connectivity information throughout the encoding–decoding process. By mapping different realizations of a system — generated from the same underlying parameters — into a continuous, structured latent space, GAUDI disentangles invariant process-level features from stochastic noise. We demonstrate its power across multiple applications, including modeling small-world networks, characterizing protein assemblies from super-resolution microscopy, analyzing collective motion in the Vicsek model, and capturing age-related changes in brain connectivity. This approach not only improves the analysis of complex graphs but also provides new insights into emergent phenomena across diverse scientific domains.
Youtube: Global graph features unveiled by unsupervised geometric deep learning