Antonio Ciarlo, Martin Selin, Giuseppe Pesce, Carlos Bustamante, and Giovanni Volpe
Date: 5 August 2025
Time: 9:45 AM – 10:15 AM
Place: Conv. Ctr. Room 4
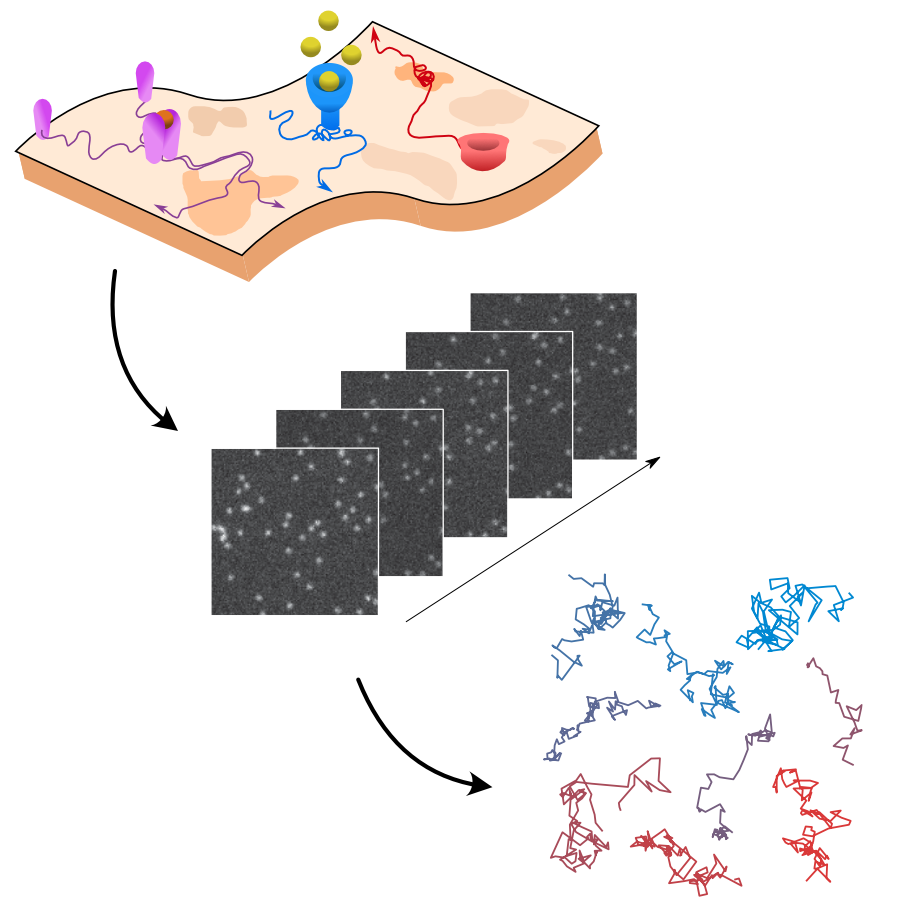
Optical tweezers are essential for single-molecule studies, providing direct access to the forces underlying biological processes such as protein folding, DNA transcription, and replication. However, manual experiments are labor-intensive, costly, and slow, especially when large data sets are required. Here we present the miniTweezer, a fully autonomous force spectroscopy platform that integrates optical tweezers with real-time image analysis and adaptive control. Once configured, it operates independently to perform high-throughput trapping, molecular attachment, and force measurements. Its robust design allows for extended unattended operation, significantly increasing the efficiency of data acquisition. We demonstrate its capabilities through DNA pulling experiments and highlight its broader applicability to microparticle interactions, colloidal assembly, and soft matter mechanics. By automating force spectroscopy, the miniTweezer enables large-scale, high-precision studies in biophysics, materials science, and nanotechnology.