
Alex is a master student in Physics at the University of Gothenburg.
During His time at the Soft Matter Lab, he will work on simulating colloidal systems with phoretic interactions.

In this work, Harshith presented his recent work on combining holographic microscopy and deep learning to study the marine microplankton. He demonstrated that the combination of holographic microscopy and deep learning can be used to follow the marine microorganisms throughout their lifespan, continuously measuring their three-dimensional positions and dry mass. The deep-learning algorithms circumvent the computationally intensive processing of holographic data and allow rapid measurements over extended periods of time. This enables to reliably estimate growth rates, both in terms of dry mass increase and cell divisions, as well as to measure trophic interactions between species such as predation events. Studying individual interactions in idealized small systems provides insights that help us understand microbial food webs and ultimately larger-scale processes. He exemplified this by showing detailed descriptions of micro-zooplankton feeding events, cell divisions, and long-term monitoring of single cells from division to division.
The article related to this presentation can be found at the following link: Microplankton life histories revealed by holographic microscopy and deep learning.



Jesús Pineda
Date: 23 August 2023
Time: 2:30 PM PDT
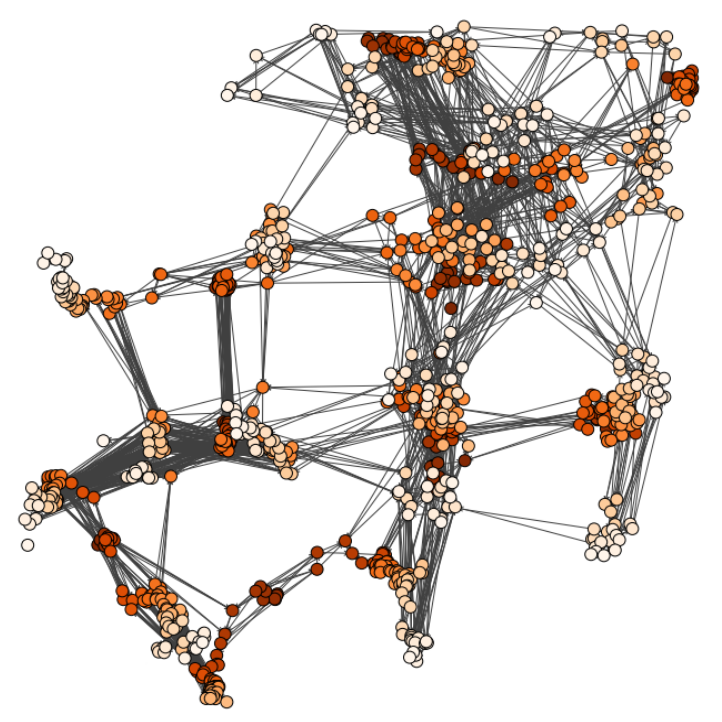
Characterizing dynamic processes in living systems provides essential information for advancing our understanding of life processes in health and diseases and for developing new technologies and treatments. In the past two decades, optical microscopy has undergone significant developments, enabling us to study the motion of cells, organelles, and individual molecules with unprecedented detail at various scales in space and time. However, analyzing the dynamic processes that occur in complex and crowded environments remains a challenge. This work introduces MAGIK, a deep-learning framework for the analysis of biological system dynamics from time-lapse microscopy. MAGIK models the movement and interactions of particles through a directed graph where nodes represent detections and edges connect spatiotemporally close nodes. The framework utilizes an attention-based graph neural network (GNN) to process the graph and modulate the strength of associations between its elements, enabling MAGIK to derive insights into the dynamics of the systems. MAGIK provides a key enabling technology to estimate any dynamic aspect of the particles, from reconstructing their trajectories to inferring local and global dynamics. We demonstrate the flexibility and reliability of the framework by applying it to real and simulated data corresponding to a broad range of biological experiments.
Reference
Pineda, J., Midtvedt, B., Bachimanchi, H. et al. Geometric deep learning reveals the spatiotemporal features of microscopic motion. Nat Mach Intell 5, 71–82 (2023)
Benjamin Midtvedt, Jesus Pineda, Fredrik Skärberg, Erik Olsén, Harshith Bachimanchi, Emelie Wesén, Elin Esbjörner, Erik Selander, Fredrik Höök, Daniel Midtvedt, Giovanni Volpe
Date: 23 August 2023
Time: 10:30 AM (PDT)
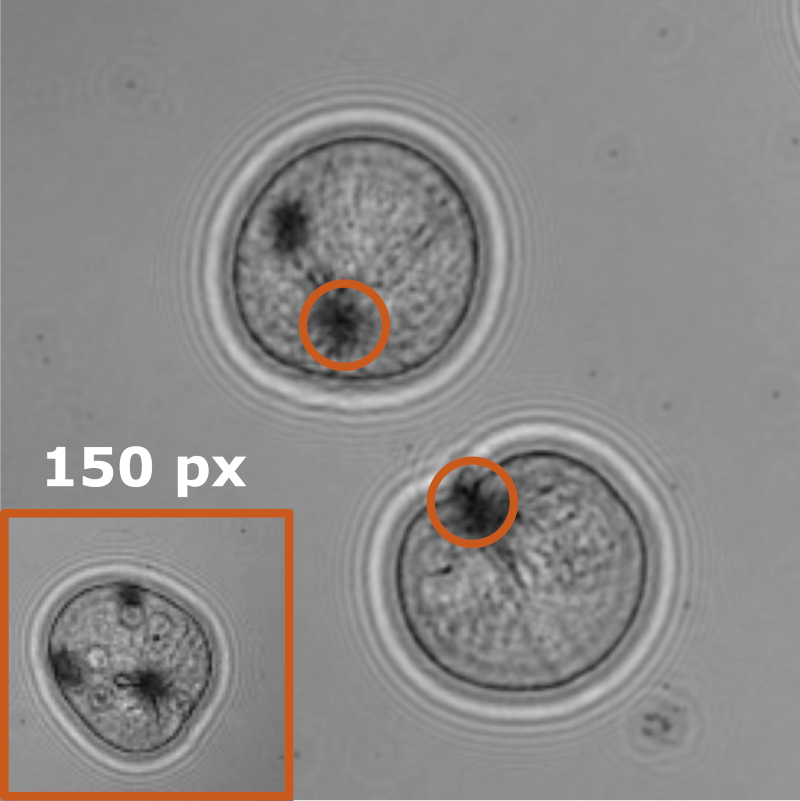
Object detection is a fundamental task in digital microscopy. Recently, machine-learning approaches have made great strides in overcoming the limitations of more classical approaches. The training of state-of-the-art machine-learning methods almost universally relies on either vast amounts of labeled experimental data or the ability to numerically simulate realistic datasets. However, the data produced by experiments are often challenging to label and cannot be easily reproduced numerically. Here, we propose a novel deep-learning method, named LodeSTAR (Low-shot deep Symmetric Tracking And Regression), that learns to detect small, spatially confined, and largely homogeneous objects that have sufficient contrast to the background with sub-pixel accuracy from a single unlabeled experimental image. This is made possible by exploiting the inherent roto-translational symmetries of the data. We demonstrate that LodeSTAR outperforms traditional methods in terms of accuracy. Furthermore, we analyze challenging experimental data containing densely packed cells or noisy backgrounds. We also exploit additional symmetries to extend the measurable particle properties to the particle’s vertical position by propagating the signal in Fourier space and its polarizability by scaling the signal strength. Thanks to the ability to train deep-learning models with a single unlabeled image, LodeSTAR can accelerate the development of high-quality microscopic analysis pipelines for engineering, biology, and medicine.
Henrik Klein Moberg
Date: 23 August 2023
Time: 8:15 AM PDT
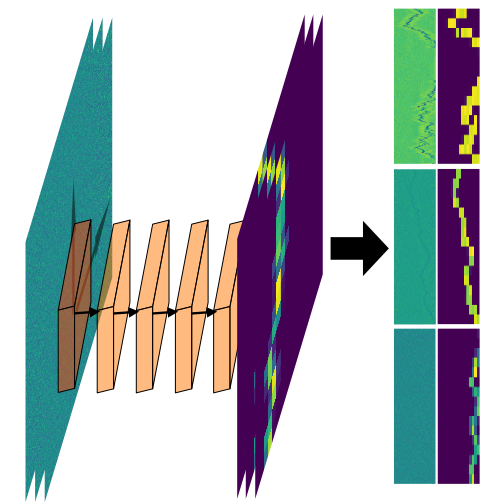
We show that a custom ResNet-inspired CNN architecture trained on simulated biomolecule trajectories surpasses the performance of standard algorithms in terms of tracking and determining the molecular weight and hydrodynamic radius of biomolecules in the low-kDa regime in optical microscopy. We show that high accuracy and precision is retained even below the 10-kDa regime, constituting approximately an order of magnitude improvement in limit of detection compared to current state-of-the-art, enabling analysis of hitherto elusive species of biomolecules such as cytokines (~5-25 kDa) important for cancer research and the protein hormone insulin (~5.6 kDa), potentially opening up entirely new avenues of biological research.
Giovanni Volpe
Date: 23 August 2023
Time: 8:00 AM PDT
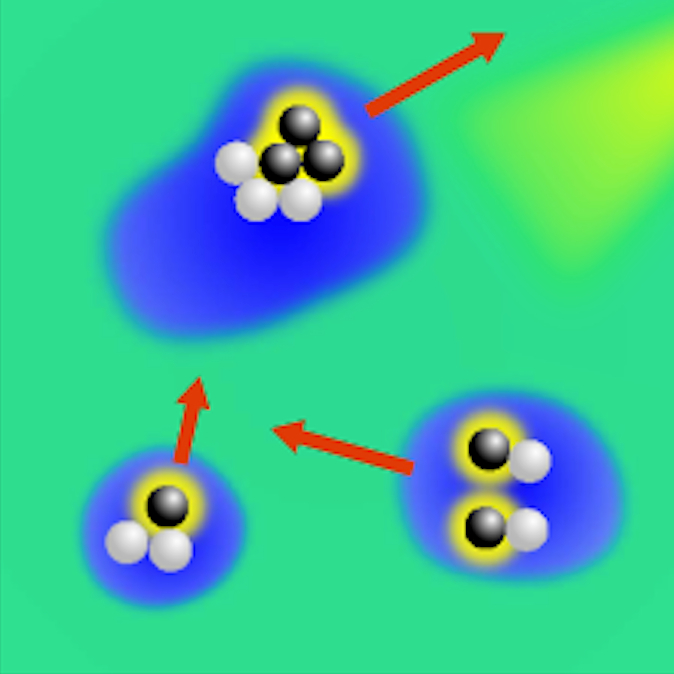
Critical Casimir forces (CCF) are a powerful tool to control the self-assembly and complex behavior of microscopic and nanoscopic colloids. While CCF were theoretically predicted in 1978, their first direct experimental evidence was provided only in 2008, using total internal reflection microscopy (TIRM). Since then, these forces have been investigated under various conditions, for example, by varying the properties of the involved surfaces or with moving boundaries. In addition, a number of studies of the phase behavior of colloidal dispersions in a critical mixture indicate critical Casimir forces as candidates for tuning the self-assembly of nanostructures and quantum dots, while analogous fluctuation-induced effects have been investigated, for example, at the percolation transition of a chemical sol, in the presence of temperature gradients, and even in granular fluids and active matter. In this presentation, I’ll give an overview of this field with a focus on recent results on the measurement of many-body forces in critical Casimir forces, the realization of micro- and nanoscopic engines powered by critical fluctuations, and the creation of light-controllable colloidal molecules and active droploids.

Falko Schmidt, Agnese Callegari, Abdallah Daddi-Moussa-Ider, Battulga Munkhbat, Ruggero Verre, Timur Shegai, Mikael Käll, Hartmut Löwen, Andrea Gambassi and Giovanni Volpe
SPIE-MNM, San Diego, CA, USA, 20 – 24 August 2023
Date: 23 August 2023
Casimir forces in quantum electrodynamics emerge between microscopic metallic objects because of the confinement of the vacuum electromagnetic fluctuations occurring even at zero temperature. Their generalization at finite temperature and in material media are referred to as Casimir–Lifshitz forces. These forces are typically attractive, leading to the widespread problem of stiction between the metallic parts of micro- and nanodevices. Recently, repulsive Casimir forces have been experimentally realized but their reliance on specialized materials prevents their dynamic control and thus limits their further applicability. Here, we experimentally demonstrate that repulsive critical Casimir forces, which emerge in a critical binary liquid mixture upon approaching the critical temperature, can be used to actively control microscopic and nanoscopic objects with nanometer precision. We demonstrate this by using critical Casimir forces to prevent the stiction caused by the Casimir–Lifshitz forces. We study a microscopic gold flake above a flat gold-coated substrate immersed in a critical mixture. Far from the critical temperature, stiction occurs because of dominant Casimir–Lifshitz forces. Upon approaching the critical temperature, we observe the emergence of repulsive critical Casimir forces that are sufficiently strong to counteract stiction. Our method provides a novel way of controlling the distances of micro- and nanostructures using tunable critical Casimir forces to counteract forces such as the Casimir–Lifshitz force, thereby preventing stiction and device failure. Due to the simplicity of our design the concept can be adapted to already existing MEMS and NEMS by, for example, controlling the temperature via light illumination.
Reference
Falko Schmidt, Agnese Callegari, Abdallah Daddi-Moussa-Ider, Battulga Munkhbat, Ruggero Verre, Timur Shegai, Mikael Käll, Hartmut Löwen, Andrea Gambassi and Giovanni Volpe, Tunable critical Casimir forces counteract Casimir-Lifshitz attraction, Nature Physics 19, 271-278 (2023)
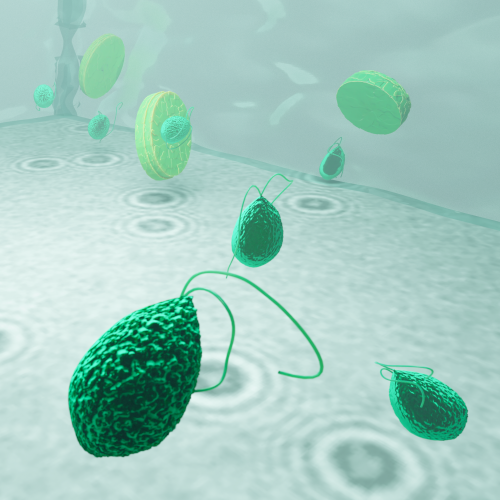
Harshith Bachimanchi
23 August 2023, 8:45 AM PDT
The marine microbial food web plays a central role in the global carbon cycle. However, our mechanistic understanding of the ocean is biased toward its larger constituents, while rates and biomass fluxes in the microbial food web are mainly inferred from indirect measurements and ensemble averages. Yet, resolution at the level of the individual microplankton is required to advance our understanding of the microbial food web. Here, we demonstrate that, by combining holographic microscopy with deep learning, we can follow microplanktons throughout their lifespan, continuously measuring their three-dimensional position and dry mass. The deep-learning algorithms circumvent the computationally intensive processing of holographic data and allow rapid measurements over extended time periods. This permits us to reliably estimate growth rates, both in terms of dry mass increase and cell divisions, as well as to measure trophic interactions between species such as predation events. The individual resolution provides information about selectivity, individual feeding rates, and handling times for individual microplanktons. The method is particularly useful to detail the rates and routes of organic matter transfer in micro-zooplankton, the most important and least known group of primary consumers in the oceans. Studying individual interactions in idealized small systems provides insights that help us understand microbial food webs and ultimately larger-scale processes. We exemplify this by detailed descriptions of micro-zooplankton feeding events, cell divisions, and long-term monitoring of single cells from division to division.
The article related to this presentation can be found at the following link: Microplankton life histories revealed by holographic microscopy and deep learning.
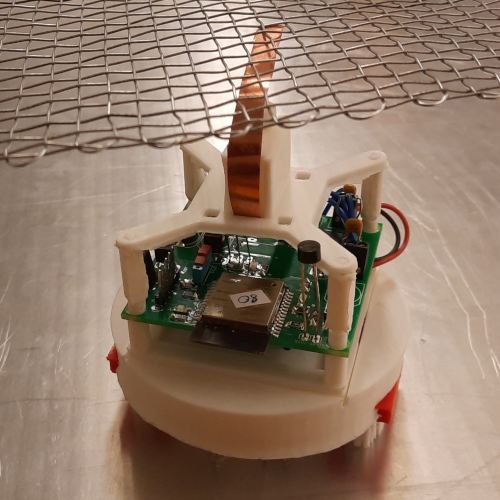
Laura Natali
Date: 22 August 2023
Time: 5:30 PM PDT
Artificial neural networks have limitations compared to biological counterparts as the latter can dynamically change connections and recover from damage. To simplify the study of connections evolving over time, we propose using programmable robots as a swarm to perform supervised learning and autonomously restructure the network. This experimental setup offers a way to study the evolution of connections in a simplified system while addressing the complexity of biological neurons. It has the potential to yield insights into the functioning of biological neural networks while providing a practical application in solving tasks.
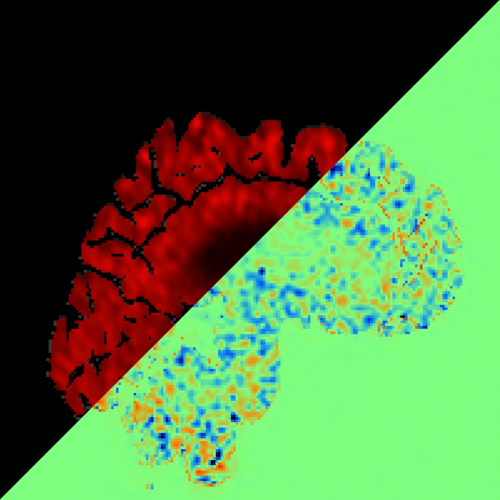
Yu-Wei Chang, Giovanni Volpe, Joana B Pereira
Date: 22 August 2023
Time: 10:15 AM PDT
In vivo tau-positron emission tomography (PET) is crucial for determining the stage of Alzheimer’s disease (AD). However, this method is expensive, not widely available, and exposes patients to ionizing radiation, which poses a carcinogenic risk. To address this issue, I’ll present our proposed method, a deep-learning synthesis approach for follow-up tau-PET brain images from baseline tau-PET images using a generative adversarial network (GAN). This technique has the potential to provide valuable insights into the progression of AD, the effectiveness of new treatments, and more accurate diagnosis of the disease.