
Sergi is a PhD student at the Biomedicine Department of the University of Vic, Cataluña, Spain.
During his time at the Soft Matter Lab, he will be working on a biophysical application of deep learning.
He will stay in our lab till 2 June 2023.

Sergi is a PhD student at the Biomedicine Department of the University of Vic, Cataluña, Spain.
During his time at the Soft Matter Lab, he will be working on a biophysical application of deep learning.
He will stay in our lab till 2 June 2023.

Christian is a master student in Complex Adaptive Systems at Chalmers University of Technology.
During his time at the Soft Matter Lab, he will study the characterization of active matter particle systems with graph neural networks.
Natsuko Rivera-Yoshida
19 January 2023
16:30, Nexus
Physical environment contribute to both the robustness and the variation of developmental trajectories and, eventually, to the evolutionary transitions. But how? Myxococcus xanthus is a soil bacterium and is widely used as a biological model. In starvation conditions, cells move individually over the substrate into growing groups of cells which, eventually, organize into three-dimensional structures called fruiting bodies. Commonly, this developmental process is studied using standard experimental protocols that employ homogeneous and flat agar substrates, without considering ecologically relevant variables. However M. Xanthus has shown to drastically alter its development when modifying variables such as the substrate topography or stiffness. This modifications occur with trait and scale specificity, at the level of individual cells, large group of cells, fruiting bodies and also at the population scale. We use experimental and analytical tools to study how multicellular organization is altered at different spatial scales and developmental moments.
Individual and collective motion of nematic, polar, and chiral actively driven objects
Andreas Menzel
19 January 2023
15:30, Nexus
Abstract:
Actively driven objects comprise a manifold of possible different realizations: from self-propelling bacteria and artificial phoretically driven colloidal particles via vibrated hoppers to walking pedestrians. We analyze basic theoretical models to identify generic features of subclasses of such agents. Within this framework, we first address nematic objects [1]. They predominantly propel along one specific axis of their body, but do not feature an explicit head or tail. That is, they can move either way by spontaneous symmetry breaking. This leads to characteristic kinks along their trajectories. Second, we study chiral objects that show persistent bending of their trajectories and migrate in discrete steps [2]. When, additionally, they tend to migrate towards a fixed remote target, rich nonlinear dynamics emerges. It comprises period doubling and chaotic behavior as a function of the tendency of alignment, which is reflected by the trajectories. Third, we consider the collective motion of continuously moving chiral objects in crystal-like arrangements [3]. We here identify a localization transition with increasing chirality or self-shearing phenomena within the crystal-like structures. Overall, we hope by our work to stimulate experimental realization and observation of the various investigated systems and phenomena.
References
[1] A. M. Menzel, J. Chem. Phys. 157, 011102 (2022).
[2] A. M. Menzel, resubmitted.
[3] Z.-F. Huang, A. M. Menzel, H. Löwen, Phys. Rev. Lett. 125, 218002 (2020).
Short Bio:
Andreas Menzel studied physics at the University of Bayreuth (Germany), where he also completed his PhD on the continuum theory of soft elastic liquid-crystalline composite materials. After postdoctoral stays at the University of Illinois at Urbana-Champaign with Prof. Nigel Goldenfeld and at the Max Planck Institute for Polymer Research in Mainz in the department headed by Prof. Kurt Kremer, as well as research stays at Kyoto University with Prof. Takao Ohta, he completed his Habilitation at Heinrich Heine University Düsseldorf at the Theory Institute for Soft Matter headed by Prof. Hartmut Löwen. Amongst others, Andreas is interested in developing and applying explicit Green’s functions methods, statistical descriptions, and continuum theories on soft matter, addressing, for example, functionalized elastic composite materials and active matter. In 2020 he moved as a Heisenberg Fellow of the German Research Foundation to Otto von Guericke University Magdeburg (Germany), where he now heads the department on Theory of Soft Matter / Biophysics.
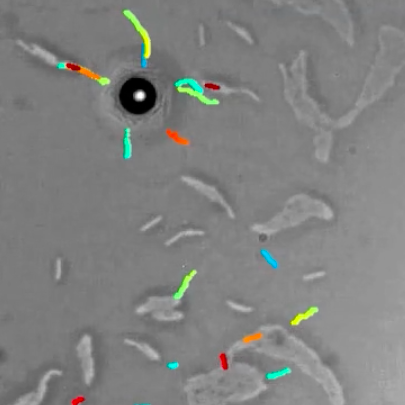
Jesús Pineda, Benjamin Midtvedt, Harshith Bachimanchi, Sergio Noé, Daniel Midtvedt, Giovanni Volpe, Carlo Manzo
Nature Machine Intelligence 5, 71–82 (2023)
arXiv: 2202.06355
doi: 10.1038/s42256-022-00595-0
The characterization of dynamical processes in living systems provides important clues for their mechanistic interpretation and link to biological functions. Thanks to recent advances in microscopy techniques, it is now possible to routinely record the motion of cells, organelles, and individual molecules at multiple spatiotemporal scales in physiological conditions. However, the automated analysis of dynamics occurring in crowded and complex environments still lags behind the acquisition of microscopic image sequences. Here, we present a framework based on geometric deep learning that achieves the accurate estimation of dynamical properties in various biologically-relevant scenarios. This deep-learning approach relies on a graph neural network enhanced by attention-based components. By processing object features with geometric priors, the network is capable of performing multiple tasks, from linking coordinates into trajectories to inferring local and global dynamic properties. We demonstrate the flexibility and reliability of this approach by applying it to real and simulated data corresponding to a broad range of biological experiments.
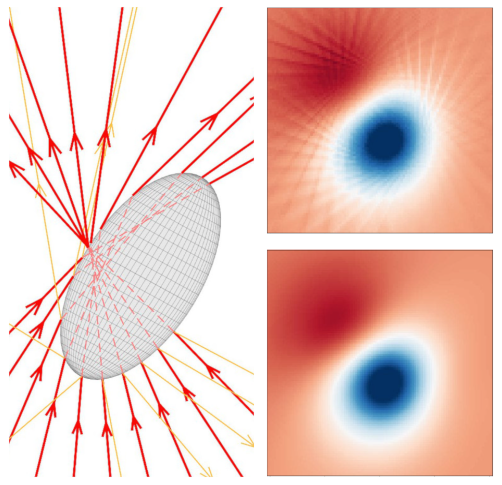
David Bronte Ciriza, Alessandro Magazzù, Agnese Callegari, Gunther Barbosa, Antonio A. R. Neves, Maria A. Iatì, Giovanni Volpe, Onofrio M. Maragò
ACS Photonics 10, 234–241 (2023)
doi: 10.1021/acsphotonics.2c01565
arXiv: 2209.04032
Optical forces are often calculated by discretizing the trapping light beam into a set of rays and using geometrical optics to compute the exchange of momentum. However, the number of rays sets a trade-off between calculation speed and accuracy. Here, we show that using neural networks permits one to overcome this limitation, obtaining not only faster but also more accurate simulations. We demonstrate this using an optically trapped spherical particle for which we obtain an analytical solution to use as ground truth. Then, we take advantage of the acceleration provided by neural networks to study the dynamics of an ellipsoidal particle in a double trap, which would be computationally impossible otherwise.

Juan S. Sierra, Jesus Pineda, Daniela Rueda, Alejandro Tello, Angelica M. Prada, Virgilio Galvis, Giovanni Volpe, Maria S. Millan, Lenny A. Romero, Andres G. Marrugo
Biomedical Optics Express 14, 335-351 (2023)
doi: 10.1364/BOE.477495
arXiv: 2210.07102
Specular microscopy assessment of the human corneal endothelium (CE) in Fuchs’ dystrophy is challenging due to the presence of dark image regions called guttae. This paper proposes a UNet-based segmentation approach that requires minimal post-processing and achieves reliable CE morphometric assessment and guttae identification across all degrees of Fuchs’ dystrophy. We cast the segmentation problem as a regression task of the cell and gutta signed distance maps instead of a pixel-level classification task as typically done with UNets. Compared to the conventional UNet classification approach, the distance-map regression approach converges faster in clinically relevant parameters. It also produces morphometric parameters that agree with the manually-segmented ground-truth data, namely the average cell density difference of -41.9 cells/mm2 (95% confidence interval (CI) [-306.2, 222.5]) and the average difference of mean cell area of 14.8 um2 (95% CI [-41.9, 71.5]). These results suggest a promising alternative for CE assessment.

In the seminar, Marcel Rey talked about his recent advances on understanding the behaviour of stimuli-responsive emulsions and afterwards introduced a simple yet versatile strategy to overcome the coffee ring effect and obtain homogeneous drying of particle dispersions.
Temperature-responsive emulsions combine the long-term stability with controlled on-demand release of the encapsulated liquid. The destabilization has previously been attributed to microgel shrinkage, leading to a lower surface coverage which induces coalescence. We demonstrated that breaking mechanism is fundamentally different than previously thought. Breaking only occurs if the stabilizing soft microgel particles assume a characteristic double-corona microstructure, which serve as weak link enabling stimuli-responsive emulsion behavior. Conversely, emulsions stabilized by regular single-corona microgels remain remarkably insensitive to temperature.
After spilling coffee, a tell-tale circular stain is left by the drying droplet. This universal phenomenon, known as the “coffee ring effect”, is observed independent of the suspended material. We recently developed a simple yet versatile strategy to achieve homogeneous drying of dispersed particles. Modifying the particle surface with surface-active polymers provides enhanced steric stabilization and facilitates adsorption to the liquid/air interface which, after drying, leads to uniform particle deposition. This method is independent of particle size and shape and applicable to a variety of commercial pigment particles promising applications in daily life.
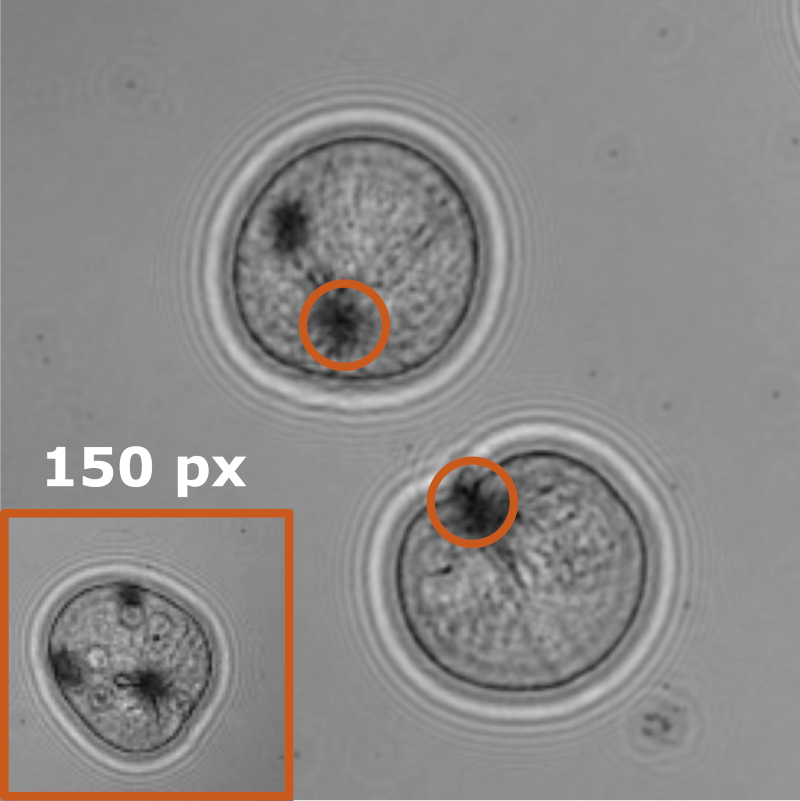
Benjamin Midtvedt, Jesús Pineda, Fredrik Skärberg, Erik Olsén, Harshith Bachimanchi, Emelie Wesén, Elin K. Esbjörner, Erik Selander, Fredrik Höök, Daniel Midtvedt, Giovanni Volpe
Nature Communications 13, 7492 (2022)
arXiv: 2202.13546
doi: 10.1038/s41467-022-35004-y
Object detection is a fundamental task in digital microscopy, where machine learning has made great strides in overcoming the limitations of classical approaches. The training of state-of-the-art machine-learning methods almost universally relies on vast amounts of labeled experimental data or the ability to numerically simulate realistic datasets. However, experimental data are often challenging to label and cannot be easily reproduced numerically. Here, we propose a deep-learning method, named LodeSTAR (Localization and detection from Symmetries, Translations And Rotations), that learns to detect microscopic objects with sub-pixel accuracy from a single unlabeled experimental image by exploiting the inherent roto-translational symmetries of this task. We demonstrate that LodeSTAR outperforms traditional methods in terms of accuracy, also when analyzing challenging experimental data containing densely packed cells or noisy backgrounds. Furthermore, by exploiting additional symmetries we show that LodeSTAR can measure other properties, e.g., vertical position and polarizability in holographic microscopy.