
Yihuai is a master student in Data Science and AI at Chalmers University of Technology.
During his time at the Soft Matter lab, he will be working on developing reinforcement learning algorithm for robotic applications.

Matilda is a master student in Engineering Physics at Chalmers University of Technology.
During her time at the Soft Matter Lab, she will be working on developing self-supervised deep learning methods for analyzing microscopy data.
Giorgio Volpe, Nuno A M Araújo, Maria Guix, Mark Miodownik, Nicolas Martin, Laura Alvarez, Juliane Simmchen, Roberto Di Leonardo, Nicola Pellicciotta, Quentin Martinet, Jérémie Palacci, Wai Kit Ng, Dhruv Saxena, Riccardo Sapienza, Sara Nadine, João F Mano, Reza Mahdavi, Caroline Beck Adiels, Joe Forth, Christian Santangelo, Stefano Palagi, Ji Min Seok, Victoria A Webster-Wood, Shuhong Wang, Lining Yao, Amirreza Aghakhani, Thomas Barois, Hamid Kellay, Corentin Coulais, Martin van Hecke, Christopher J Pierce, Tianyu Wang, Baxi Chong, Daniel I Goldman, Andreagiovanni Reina, Vito Trianni, Giovanni Volpe, Richard Beckett, Sean P Nair, Rachel Armstrong
Journal of Physics: Condensed Matter 37, 333501 (2025)
arXiv: 2407.10623
doi: 10.1088/1361-648X/adebd3
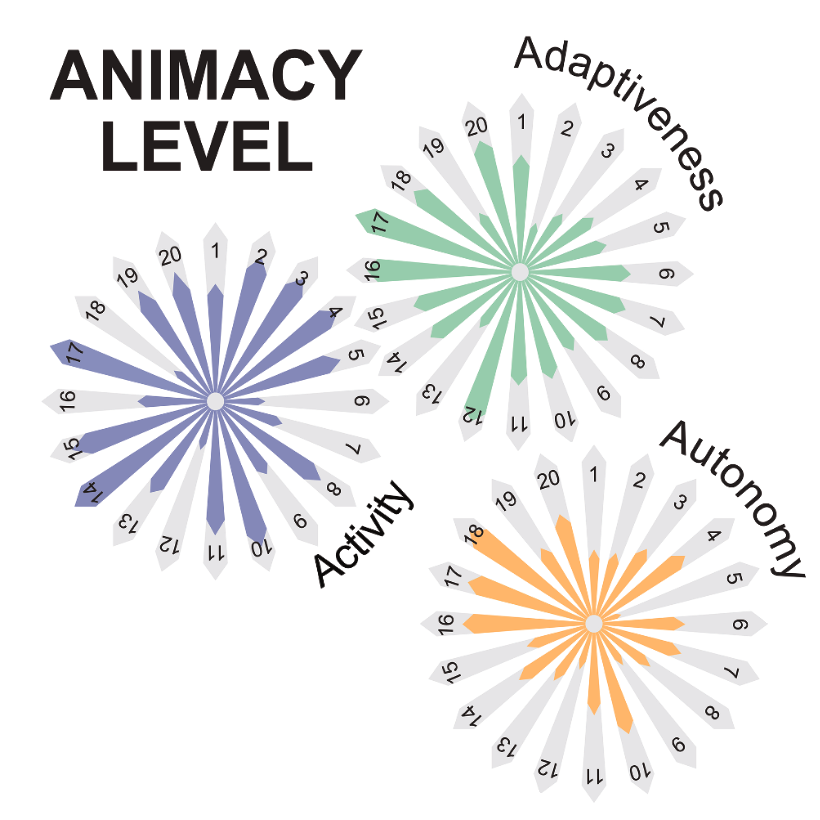
Humanity has long sought inspiration from nature to innovate materials and devices. As science advances, nature-inspired materials are becoming part of our lives. Animate materials, characterized by their activity, adaptability, and autonomy, emulate properties of living systems. While only biological materials fully embody these principles, artificial versions are advancing rapidly, promising transformative impacts in the circular economy, health and climate resilience within a generation. This roadmap presents authoritative perspectives on animate materials across different disciplines and scales, highlighting their interdisciplinary nature and potential applications in diverse fields including nanotechnology, robotics and the built environment. It underscores the need for concerted efforts to address shared challenges such as complexity management, scalability, evolvability, interdisciplinary collaboration, and ethical and environmental considerations. The framework defined by classifying materials based on their level of animacy can guide this emerging field to encourage cooperation and responsible development. By unravelling the mysteries of living matter and leveraging its principles, we can design materials and systems that will transform our world in a more sustainable manner.

Jun Yi holds a bachelor’s degree in Chemistry from the University of Münster.
During his internship at the Soft Matter Lab, he will investigate the interactions of polymer-coated silica microparticles under various stimuli using optical tweezers.
The Soft Matter Lab participates to the SPIE Optics+Photonics conference in San Diego, CA, USA, 3-7 August 2025, with the presentations listed below.
Giovanni Volpe, who serves as Symposium Chair for the SPIE Optics+Photonics Congress in 2025, is a coauthor of the following invited presentations:
Giovanni Volpe will also be the reference presenter of the following Poster contributions:

Blanca Zufiria-Gerbolés, Mite Mijalkov, Ludvig Storm, Dániel Veréb, Zhilei Xu, Anna Canal-Garcia, Jiawei Sun, Yu-Wei Chang, Hang Zhao, Emiliano Gómez-Ruiz, Massimiliano Passaretti, Sara Garcia-Ptacek, Miia Kivipelto, Per Svenningsson, Henrik Zetterberg, Heidi Jacobs, Kathy Lüdge, Daniel Brunner, Bernhard Mehlig, Giovanni Volpe, Joana B. Pereira
SPIE-ETAI, San Diego, CA, USA, 3 – 7 August 2025
Date: 7 August 2025
Time: 11:15 AM – 11:45 AM PDT
Place: Conv. Ctr. Room 4
Using reservoir computing and diffusion-weighted imaging, we explored changes in brain connectivity patterns and their impact on cognition during aging. We found that whole-brain networks perform optimally at low densities, with performance decreasing as network density increases, particularly in regions with weaker connections. This decline was strongly associated with age and cognitive performance. Our results suggest that a core network of anatomical hubs is essential for optimal brain function, while peripheral connections are more vulnerable to aging. This study highlights the potential of reservoir computing for understanding age-related cognitive decline.
Reference
Mite Mijalkov, Ludvig Storm, Blanca Zufiria-Gerbolés, Dániel Veréb, Zhilei Xu, Anna Canal-Garcia, Jiawei Sun, Yu-Wei Chang, Hang Zhao, Emiliano Gómez-Ruiz, Massimiliano Passaretti, Sara Garcia-Ptacek, Miia Kivipelto, Per Svenningsson, Henrik Zetterberg, Heidi Jacobs, Kathy Lüdge, Daniel Brunner, Bernhard Mehlig, Giovanni Volpe, Joana B. Pereira, Computational memory capacity predicts aging and cognitive decline
Nature Communications 16, 2748 (2025)

Anoop C. Patil, Benny Jian Rong Sng, Yu-Wei Chang, Joana B. Pereira, Chua Nam-Hai, Rajani Sarojam, Gajendra Pratap Singh, In-Cheol Jang, and Giovanni Volpe
Date: 6 August 2025
Time: 10:30 AM – 11:00 AM
Place: Conv. Ctr. Room 4
Plants experience a wide variety of stresses, from light and temperature fluctuations to bacterial infections. Each stress has a biomolecular fingerprint, but detecting and interpreting these signatures across species can be challenging. This work presents a deep learning-based approach using Variational Autoencoders (VAEs) to uncover how plants respond to light stress, shade avoidance, temperature stress, and bacterial infection — all without requiring any human intervention in spectral processing. By encoding Raman spectral data into an intuitive latent space, this method automatically categorizes and visualizes stress-specific biomolecular shifts, offering a powerful, unsupervised tool for stress phenotyping in crops.
Reference:
Patil, A.C. et al. Deep-Learning Investigation of Vibrational Raman Spectra for Plant-Stress Analysis. arXiv preprint arXiv:2507.15772v1 (2025). URL https://arxiv.org/abs/2507.15772

Martin Selin, A. Ciarlo, G. Pesce, L. Bengtsson, J. Camuñas-Soler, V. Sundar Rajan, F. Westerlund, L. M. Wilhelmsson, I. Pastor, F. Ritort, S. B. Smith, C. Bustamante, G. Volpe
Date: 5 August 2025
Time: 4:30 PM – 4:45 PM PDT
Place: Conv. Ctr. Room 4
Single-molecule studies are vital for elucidating fundamental biological processes, including protein folding, DNA transcription, and replication. However, performing these experiments manually on individual molecules is notoriously time-consuming and costly. To address this challenge, we have developed a fully autonomous single-molecule force spectroscopy platform by integrating a custom-built optical tweezers instrument with real-time deep-learning-based image analysis and adaptive control protocols. Our system achieves human-level throughput in terms of experiments per hour while remaining robust enough to operate continuously for hours without intervention. We demonstrate the versatility of our platform by having it perform DNA pulling experiments on both lambda DNA and DNA hairpins fully autonomously. These results push the boundaries of high-throughput data collection in single-molecule biophysics, paving the way for merging single-molecule studies with large-scale, data-driven approaches—ultimately enabling new insights into the dynamic, transient states of complex biological systems.
Bohdan Yeroshenko, Henrik Klein Moberg, Leyla Beckerman, Joachim Fritzsche, David Albinsson, Barbora Špačková, Daniel Midtvedt, Giovanni Volpe, Christoph Langhammer
SPIE-ETAI, San Diego, CA, USA, 3 – 7 August 2025
Date: 5 August 2025
Time: 8:45 AM – 9:15 AM PDT
Place: Conv. Ctr. Room 4
Nanofluidic Scattering Microscopy (NSM) is a label-free characterization method that leverages the interference of light scattered by nanochannels and single molecules within them. This technique enables accurate determination of molecular weight and hydrodynamic radius based solely on scattering, without requiring prior molecular knowledge. However, standard analysis methods limit NSM’s sensitivity to 66 kDa for proteins. In this presentation, I will demonstrate how we push this detection limit an order of magnitude further by integrating ultrasmall geometry with an advanced machine learning analysis approach, all while maintaining the same input laser power intensity.
Angelo Barona Balda, Aykut Argun, Agnese Callegari, Giovanni Volpe
SPIE-OTOM, San Diego, CA, USA, 3 – 7 August 2025
Date: 4 August 2025
Time: 5:30 PM – 7:30 PM PDT
Place: Conv. Ctr. Exhibit Hall A
Presenter: Giovanni Volpe
Contribution submitted by Agnese Callegari
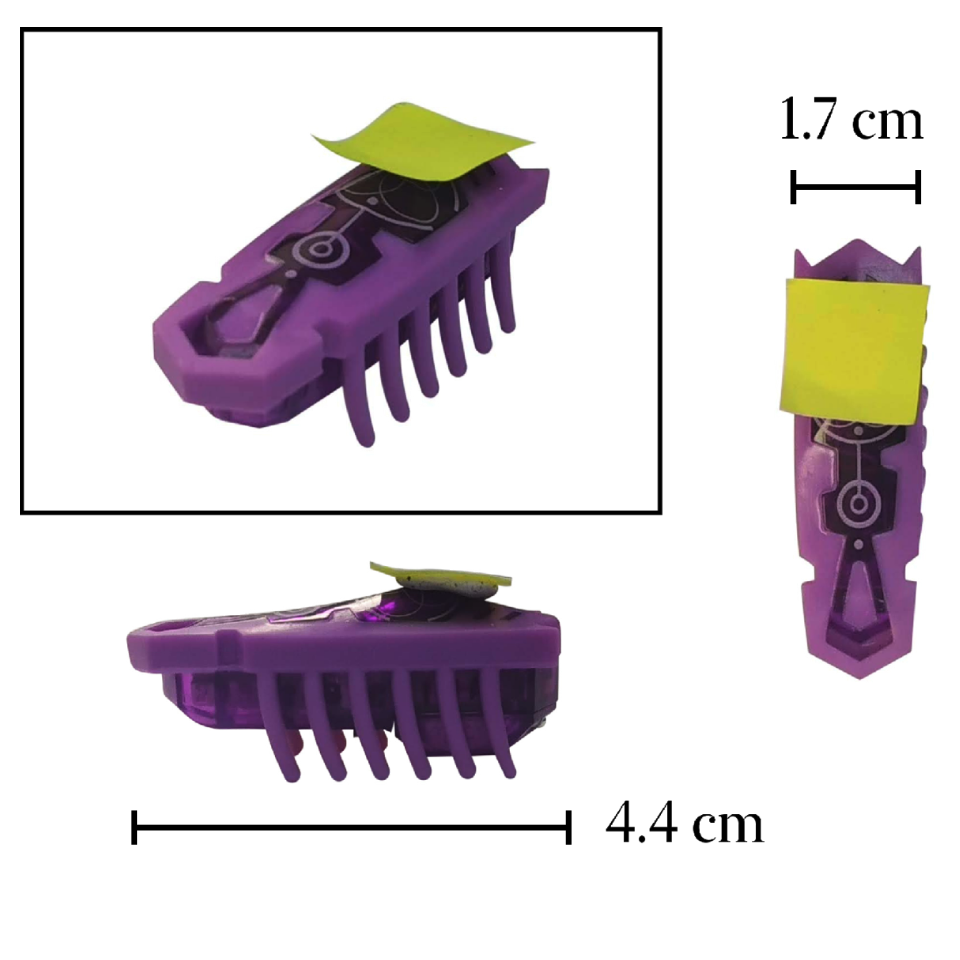
Active matter is based on concepts of nonequilibrium thermodynamics applied to the most diverse disciplines. A key concept is the active Brownian particle, which, unlike passive ones, extracts energy from its environment to generate complex motion and emergent behaviors. Despite its significance, active matter remains absent from standard curricula. This work presents macroscopic experiments using commercially available Hexbugs to demonstrate active matter phenomena. We show how Hexbugs can be modified to perform both regular and chiral active Brownian motion and interact with passive objects, inducing movement and rotation. By introducing obstacles, we sort Hexbugs based on motility and chirality. Finally, we demonstrate a Casimir-like attraction effect between planar objects in the presence of active particles.
Reference
Angelo Barona Balda, Aykut Argun, Agnese Callegari, Giovanni Volpe
Playing with Active Matter, Americal Journal of Physics 92, 847–858 (2024)