The defense took place in FB, Institutionen för fysik, Origovägen 6b, Göteborg, at 09:00.
Title: From Light to Data Using Deep Learning for Quantitative Microscopy
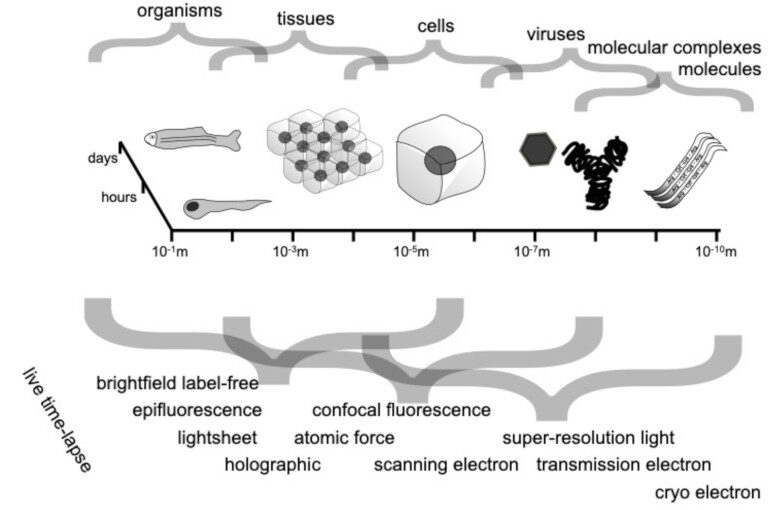
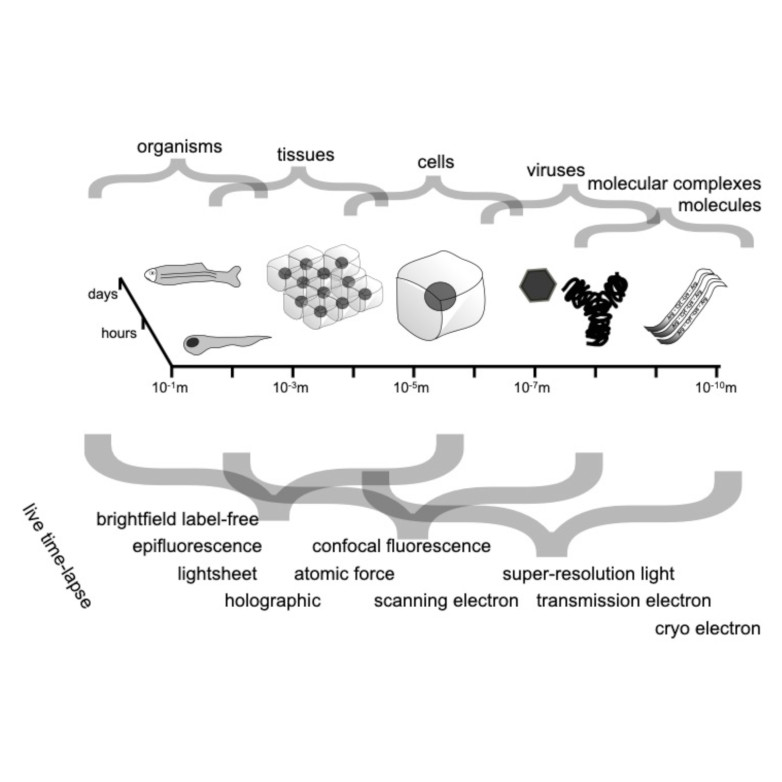
Abstract: Quantitative microscopy aims to measure physical properties of microscopic particles from optical images, but weak and complex signals often make this difficult. This thesis explores how computational methods, especially deep learning guided by physical understanding, can improve particle detection and characterization in microscopy.
The work introduces new approaches for locating and tracking particles, extends these ideas to three-dimensional and label-free imaging, and reviews practical analysis workflows. It further shows how combining complementary imaging techniques can enhance nanoparticle measurements and how deep learning can recover three-dimensional structural information from microscopy images.
Overall, this thesis strengthens the connection between optical measurements and quantitative particle information, expanding the potential of label-free microscopy for biological and nanoscale studies.
Thesis: https://gupea.ub.gu.se/handle/2077/90201
Supervisor: Daniel Midtvedt
Examiner: Raimund Feifel
Opponent: Arrate Munoz Barrutia
Committee: Per Augustsson, Jens Petersen, Rebecka Jörnsten
Alternate board member: Vitali Zhaunerchyk