
Fredrik will defend his thesis on 29 January at 09:00 in FB-salen, Institutionen för fysik, Origovägen 6, Göteborg.

Fredrik will defend his thesis on 29 January at 09:00 in FB-salen, Institutionen för fysik, Origovägen 6, Göteborg.

flexible PET sheet. (Image by H. P. Thanabalan.)
Hari Prakash Thanabalan, Lars Bengtsson, Ugo Lafont, Giovanni Volpe
arXiv: 2512.07813
Soft robots require directional control to navigate complex terrains. However, achieving such control often requires multiple actuators, which increases mechanical complexity, complicates control systems, and raises energy consumption. Here, we introduce an inchworm-inspired soft robot whose locomotion direction is controlled passively by patterned substrates. The robot employs a single rolled dielectric elastomer actuator, while groove patterns on a 3D-printed substrate guide its alignment and trajectory. Through systematic experiments, we demonstrate that varying groove angles enables precise control of locomotion direction without the need for complex actuation strategies. This groove-guided approach reduces energy consumption, simplifies robot design, and expands the applicability of bio-inspired soft robots in fields such as search and rescue, pipe inspection, and planetary exploration.

Both Andrea Schiano di Colella and Eduard Andrei Duta Costache, the two doctoral candidates based at the University of Gothenburg, participated to the event along with the other doctoral candidates of the network.
The training event consisted of a series of lectures on different topics such as small scale robots and their actuation mechanisms, theoretical aspects of the simulation of the dynamics of bodies immersed in fluids at low Reynolds numbers, working principles and applications of swarm robotics, and finally artificial intelligence and its applications to data analysis in experiments.
The event included a visit to the Swedish Algae Factory (SAFAB) and Smogenlax on the topic of circular economy, and concluded with a series of meetings between the representatives of the participating institutions, from both accademia and industry, to exchange research questions and plan secondments.
Jesus Pineda defended his PhD thesis on November 11th, 2025. Congrats!
The defense took place in SB-H7 lecture hall, Institutionen för fysik, Johanneberg Campus, Göteborg, at 9:00.
Title: Inductive Biases for Efficient Deep Learning in Microscopy
Abstract: Deep learning has become an indispensable tool for the analysis of microscopy data, yet its integration into routine research remains uneven. Several factors contribute to this gap, including the limited availability of well-annotated datasets and the high computational demands of modern architectures. Microscopy introduces further challenges, as it spans diverse modalities and scales, from proteins to tissues, producing heterogeneous data that defy standardization. Generating reliable annotations also requires expertise and time, while unequal access to high-performance computing further widens the divide between well-resourced institutions and smaller laboratories.
This dissertation argues that the prevailing paradigm of scaling models with ever-larger datasets and computational resources yields diminishing returns for microscopy. Instead, it explores the role of inductive biases as a foundation for building models that are more data-efficient, computationally accessible, and scientifically meaningful. Inductive biases are structural assumptions embedded in model design that guide learning toward patterns aligned with the underlying problem. The first part of this work examines their central role in the advancement of modern deep learning and the diverse ways they shape model behavior.
This potential is demonstrated through three case studies. First, MAGIK employs graph neural networks to analyze biological dynamics in time-lapse microscopy, uncovering local and global properties with high precision, even when trained on limited data. Next, MIRO leverages recurrent graph neural networks to process single-molecule localization datasets, improving the efficiency and reliability of clustering for variable biological structures and scales while retaining strong generalization with minimal supervision. Finally, GAUDI introduces a representation learning framework for characterizing biological systems, providing a physically meaningful representation space for interpretable and transferable analysis.
The findings presented here demonstrate that the integration of inductive biases provides a cohesive strategy to extend the reach of deep learning in the life sciences, enhancing accessibility and ensuring scientific utility under resource constraints.
Thesis: https://gupea.ub.gu.se/items/672c7946-51d6-4773-ad8c-35a3eed41499
Supervisor: Giovanni Volpe
Co-Supervisor: Carlo Manzo
Examiner: Raimund Feifel
Opponent: Anna Kreshuk
Committee: Juliette Griffié, Daniel sage, Daniel Persson
Alternate board member: Jonas Enger

Jesús Pineda, Sergi Masó-Orriols, Montse Masoliver, Joan Bertran, Mattias Goksör, Giovanni Volpe and Carlo Manzo
Nature Communications 16, 9693 (2025)
arXiv: 2412.00173
doi: 10.1038/s41467-025-65557-7
Single-molecule localization microscopy generates point clouds corresponding to fluorophore localizations. Spatial cluster identification and analysis of these point clouds are crucial for extracting insights about molecular organization. However, this task becomes challenging in the presence of localization noise, high point density, or complex biological structures. Here, we introduce MIRO (Multifunctional Integration through Relational Optimization), an algorithm that uses recurrent graph neural networks to transform the point clouds in order to improve clustering efficiency when applying conventional clustering techniques. We show that MIRO supports simultaneous processing of clusters of different shapes and at multiple scales, demonstrating improved performance across varied datasets. Our comprehensive evaluation demonstrates MIRO’s transformative potential for single-molecule localization applications, showcasing its capability to revolutionize cluster analysis and provide accurate, reliable details of molecular architecture. In addition, MIRO’s robust clustering capabilities hold promise for applications in various fields such as neuroscience, for the analysis of neural connectivity patterns, and environmental science, for studying spatial distributions of ecological data.

Jesus Manuel Antúnez Domínguez, Laura Pérez García, Natsuko Rivera-Yoshida, Jasmin Di Franco, David Steiner, Alejandro V. Arzola, Mariana Benítez, Charlotte Hamngren Blomqvist, Roberto Cerbino, Caroline Beck Adiels, Giovanni Volpe
Soft Matter 21, 8602-8623 (2025)
arXiv: 2407.18714
doi: 10.1063/5.0235449
Myxococcus xanthus is a unicellular organism known for its capacity to move and communicate, giving rise to complex collective properties, structures and behaviors. These characteristics have contributed to position M. xanthus as a valuable model organism for exploring emergent collective phenomena at the interface of biology and physics, particularly within the growing domain of active matter research. Yet, researchers frequently encounter difficulties in establishing reproducible and reliable culturing protocols. This tutorial provides a detailed and accessible guide to the culture, growth, development, and experimental sample preparation of M. xanthus. In addition, it presents several exemplary experiments that can be conducted using these samples, including motility assays, fruiting body formation, predation, and elasticotaxis—phenomena of direct relevance for active matter studies.

Andrea has a Master degree in Theoretical Physics from the University of Naples “Federico II”, Italy.
During the course of his PhD, as part of the GREENS MSCA Doctoral Network he will focus on the development of deep learning based protocols for accurate autonomous microscopy.

Ade Satria Saloka Santosa, Yu-Wei Chang, Andreas B. Dahlin, Lars Osterlund, Giovanni Volpe, Kunli Xiong
Nature 646, 1089-1095 (2025)
arXiv: 2502.03580
doi: 10.1038/s41586-025-09642-3
As demand for immersive experiences grows, displays are moving closer to the eye with smaller sizes and higher resolutions. However, shrinking pixel emitters reduce intensity, making them harder to perceive. Electronic Papers utilize ambient light for visibility, maintaining optical contrast regardless of pixel size, but cannot achieve high resolution. We show electrically tunable meta-pixels down to ~560 nm in size (>45,000 PPI) consisting of WO3 nanodiscs, allowing one-to-one pixel-photodetector mapping on the retina when the display size matches the pupil diameter, which we call Retina Electronic Paper. Our technology also supports video display (25 Hz), high reflectance (~80%), and optical contrast (~50%), which will help create the ultimate virtual reality display.
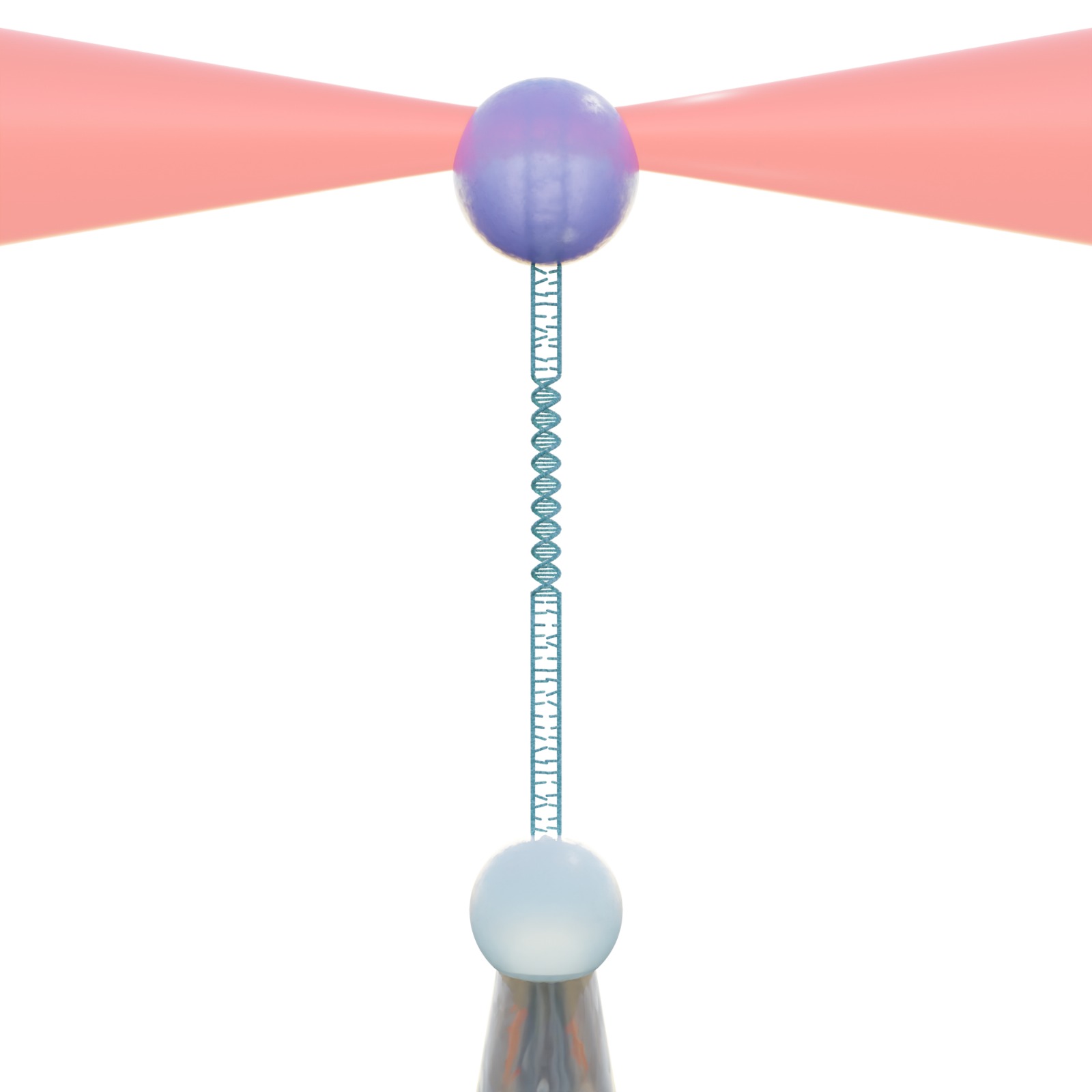
The defense took place in PJ, Institutionen för fysik, Origovägen 6b, Göteborg, at 13:00.
Title: Advanced and Autonomous Applications of Optical Tweezers
Abstract: Optical tweezers have become a central tool, using lasers to manipulate and probe objects with exceptional precision enabling single-molecule, single-cell, and single-particle studies. However, this precision comes at the cost of throughput.
By developing a fully autonomous system we can adress this limitation of optical tweezers. The system is capable of perfoming multiple different experiments independently and of operating for over 10 hours continously. Using the same system we also investigate particle adsorption into liquid-liquid interfaces revealing never before seen dynamics.
These developments help optical tweezers by bridging the gap between single-molecule, cell or particle studies and ensemble measurements, enabling the application of deep learning for advanced modeling and unlocking the potential of optical tweezers for large, data-driven studies.
Thesis: https://gupea.ub.gu.se/handle/2077/87446?show=full
Supervisor: Giovanni Volpe
Examiner: Raimund Feifel
Opponent: Borja Ibarra
Committee: Dag Hanstorp, Timo Betz, Kristine Berg-Sørensen
Alternate board member: Paolo Vinai

Prakhar has a Master’s degree in Biomedical Engineering from RWTH Aachen University in Germany.
During the course of his PhD, as part of the SPM 4.0 MSCA Doctoral Network he will focus on the development of deep learning based packages for processing of Atomic Force Microscopy data and on the development of a photonic force microscope.