Angelo Barona Balda, Aykut Argun, Agnese Callegari, Giovanni Volpe
SPIE-OTOM, San Diego, CA, USA, 18 – 22 August 2024
Date: 22 August 2024
Time: 3:00 PM – 3:15 PM
Place: Conv. Ctr. Room 6D
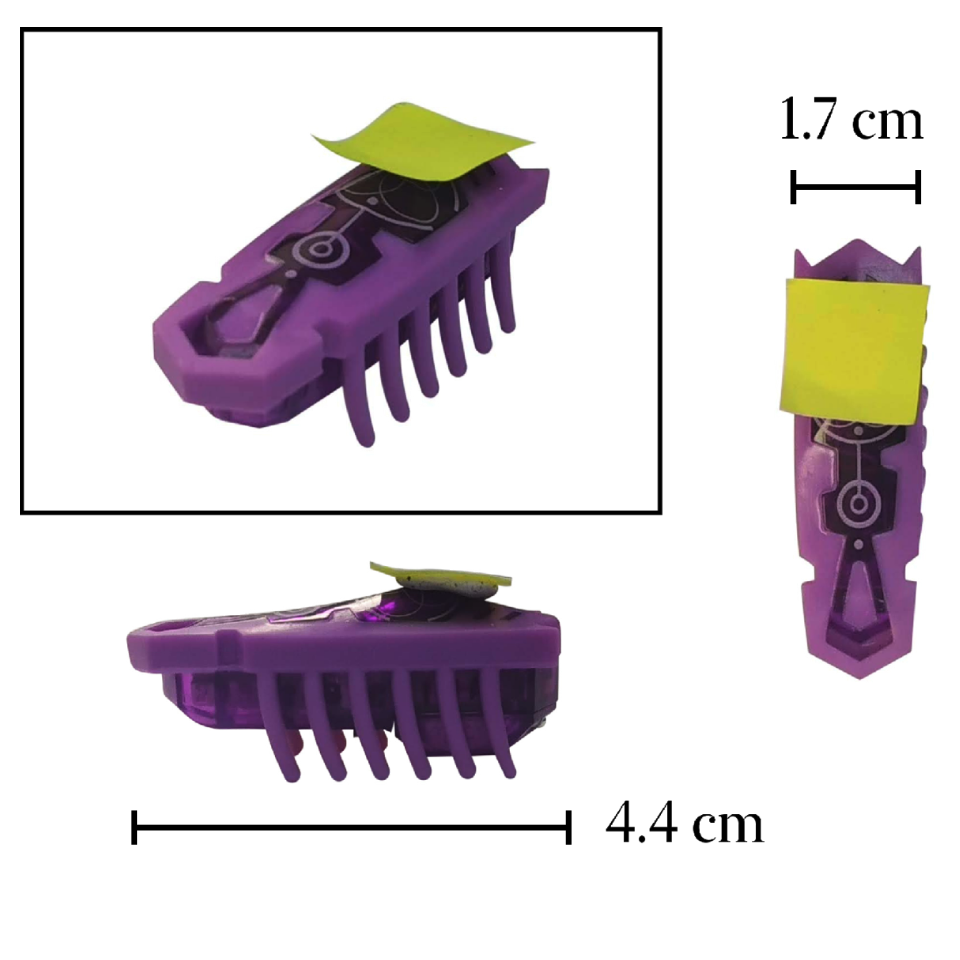
Active matter is based on concepts of nonequilibrium thermodynamics applied to the most diverse disciplines. Active Brownian particles, unlike their passive counterparts, self-propel and give rise to complex behaviors distinctive of active matter. As the field is relatively recent, active matter still lacks curricular inclusion. Here, we propose macroscopic experiments using Hexbugs, a commercial toy robot, demonstrating effects peculiar of active systems, such as the setting into motion of passive objects via active particles, the sorting of active particles based on their mobility and chirality. Additionally, we provide a demonstration of Casimir-like attraction between planar objects mediated by active particles.
Reference
Angelo Barona Balda, Aykut Argun, Agnese Callegari, Giovanni Volpe, Playing with Active Matter, arXiv: 2209.04168