Gorka Muñoz-Gil, Harshith Bachimanchi, Jesús Pineda, Benjamin Midtvedt, Gabriel Fernández-Fernández, Borja Requena, Yusef Ahsini, Solomon Asghar, Jaeyong Bae, Francisco J. Barrantes, Steen W. B. Bender, Clément Cabriel, J. Alberto Conejero, Marc Escoto, Xiaochen Feng, Rasched Haidari, Nikos S. Hatzakis, Zihan Huang, Ignacio Izeddin, Hawoong Jeong, Yuan Jiang, Jacob Kæstel-Hansen, Judith Miné-Hattab, Ran Ni, Junwoo Park, Xiang Qu, Lucas A. Saavedra, Hao Sha, Nataliya Sokolovska, Yongbing Zhang, Giorgio Volpe, Maciej Lewenstein, Ralf Metzler, Diego Krapf, Giovanni Volpe, Carlo Manzo
Nature Communications 16, 6749 (2025)
arXiv: 2311.18100
doi: https://doi.org/10.1038/s41467-025-61949-x
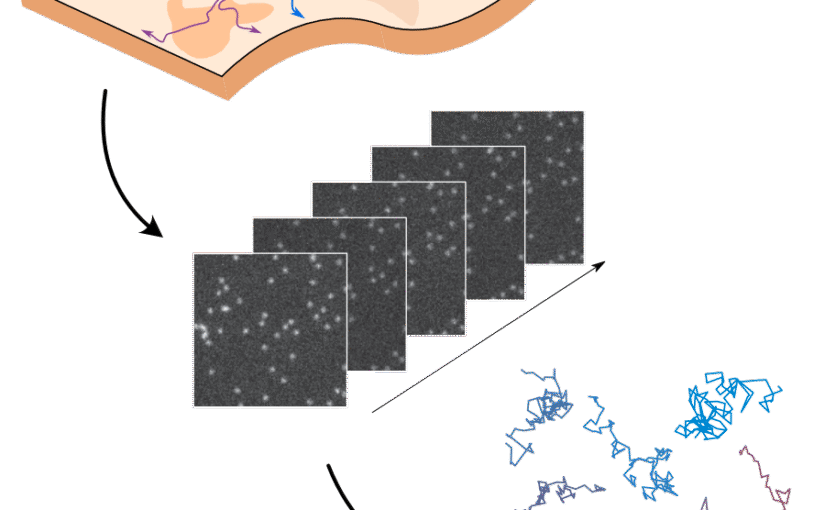
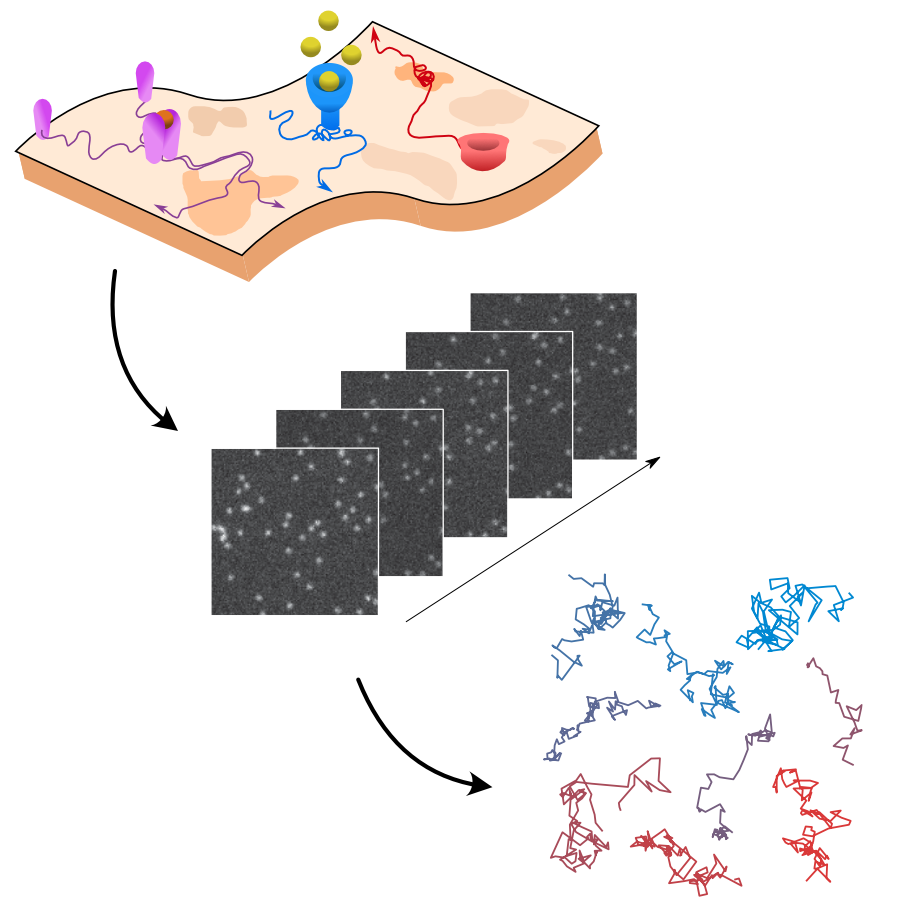
The analysis of live-cell single-molecule imaging experiments can reveal valuable information about the heterogeneity of transport processes and interactions between cell components. These characteristics are seen as motion changes in the particle trajectories. Despite the existence of multiple approaches to carry out this type of analysis, no objective assessment of these methods has been performed so far. Here, we report the results of a competition to characterize and rank the performance of these methods when analyzing the dynamic behavior of single molecules. To run this competition, we implemented a software library that simulates realistic data corresponding to widespread diffusion and interaction models, both in the form of trajectories and videos obtained in typical experimental conditions. The competition constitutes the first assessment of these methods, providing insights into the current limitations of the field, fostering the development of new approaches, and guiding researchers to identify optimal tools for analyzing their experiments.