Label-free characterization of biological matter across scales
Daniel Midtvedt, Erik Olsén, Benjamin Midtvedt, Elin K. Esbjörner, Fredrik Skärberg, Berenice Garcia, Caroline B. Adiels, Fredrik Höök, Giovanni Volpe
SPIE-ETAI
Date: 24 August 2022
Time: 09:10 (PDT)
News
Presentation by Y.-W. Chang at SPIE-ETAI, San Diego, 24 August 2022
Yu-Wei Chang, Laura Natali, Oveis Jamialahmadi, Stefano Romeo, Joana B Pereira, Giovanni Volpe
Submitted to SPIE-ETAI
Date: 24 August 2022
Time: 08:00 (PDT)
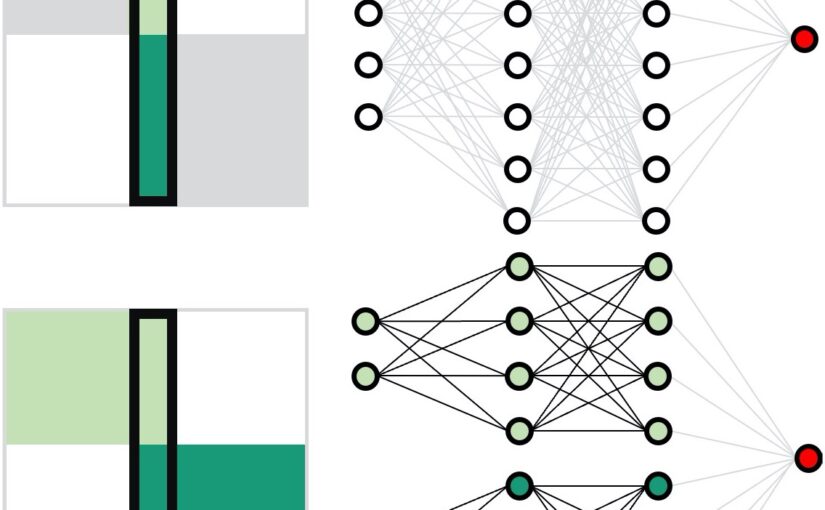
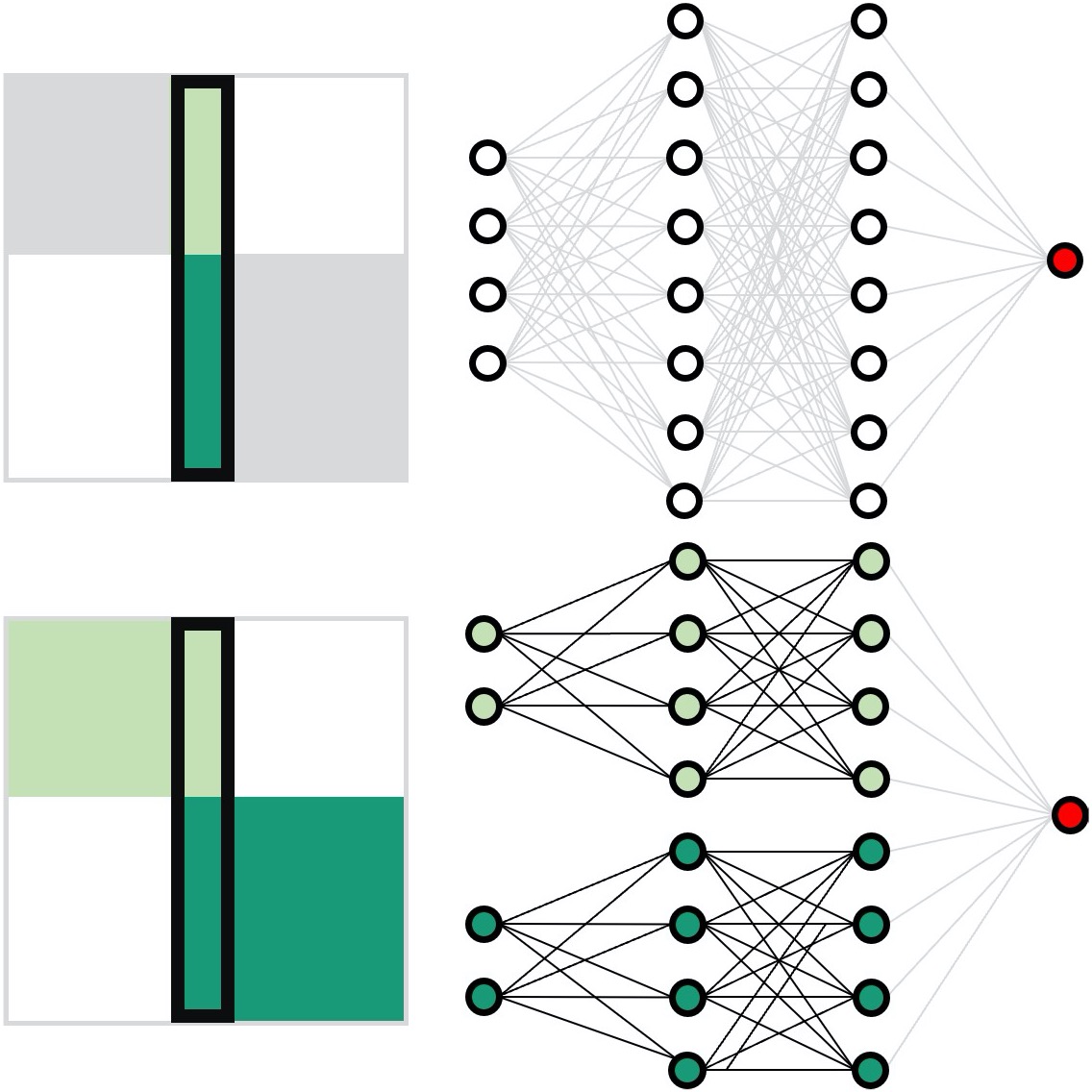
Neural network training and validation rely on the availability of large high-quality datasets. However, in many cases, only incomplete datasets are available, particularly in health care applications, where each patient typically undergoes different clinical procedures or can drop out of a study. Here, we introduce GapNet, an alternative deep-learning training approach that can use highly incomplete datasets without overfitting or introducing artefacts. Using two highly incomplete real-world medical datasets, we show that GapNet improves the identification of patients with underlying Alzheimer’s disease pathology and of patients at risk of hospitalization due to Covid-19. Compared to commonly used imputation methods, this improvement suggests that GapNet can become a general tool to handle incomplete medical datasets.
Presentation by D. Midtvedt at SPIE-ETAI, San Diego, 23 August 2022
Benjamin Midtvedt, Jesus Pineda, Fredrik Skärberg, Erik Olsén, Harshith Bachimanchi, Emelie Wesén, Elin Esbjörner, Erik Selander, Fredrik Höök, Daniel Midtvedt, Giovanni Volpe
Submitted to SPIE-ETAI
Date: 23 August 2022
Time: 2:20 PM (PDT)
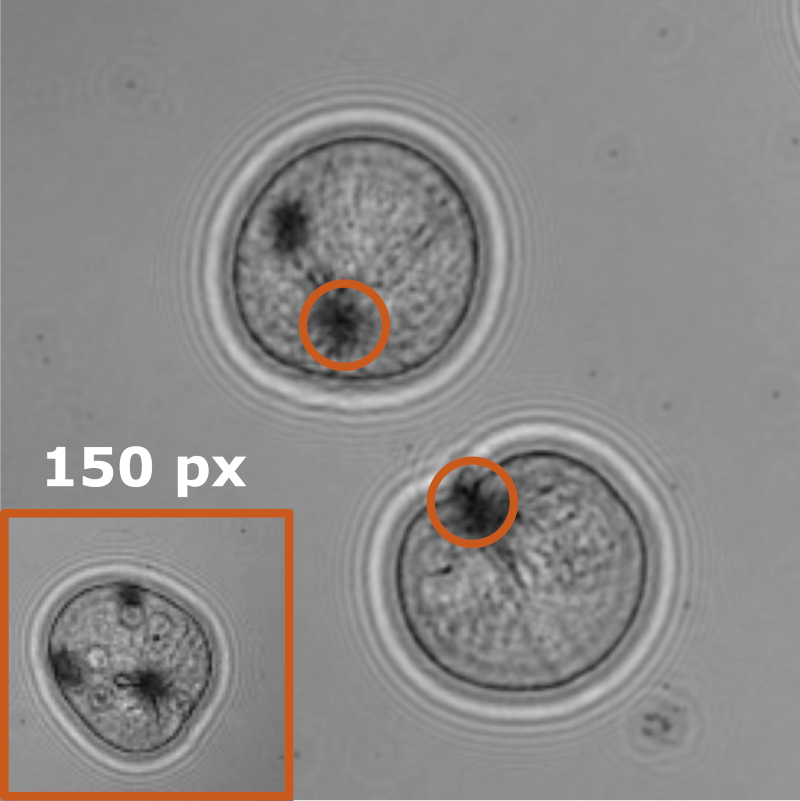
Object detection is a fundamental task in digital microscopy. Recently, machine-learning approaches have made great strides in overcoming the limitations of more classical approaches. The training of state-of-the-art machine-learning methods almost universally relies on either vast amounts of labeled experimental data or the ability to numerically simulate realistic datasets. However, the data produced by experiments are often challenging to label and cannot be easily reproduced numerically. Here, we propose a novel deep-learning method, named LodeSTAR (Low-shot deep Symmetric Tracking And Regression), that learns to detect small, spatially confined, and largely homogeneous objects that have sufficient contrast to the background with sub-pixel accuracy from a single unlabeled experimental image. This is made possible by exploiting the inherent roto-translational symmetries of the data. We demonstrate that LodeSTAR outperforms traditional methods in terms of accuracy. Furthermore, we analyze challenging experimental data containing densely packed cells or noisy backgrounds. We also exploit additional symmetries to extend the measurable particle properties to the particle’s vertical position by propagating the signal in Fourier space and its polarizability by scaling the signal strength. Thanks to the ability to train deep-learning models with a single unlabeled image, LodeSTAR can accelerate the development of high-quality microscopic analysis pipelines for engineering, biology, and medicine.
Presentation by A. Callegari at SPIE-ETAI, San Diego, 23 August 2022
Agnese Callegari, Mathias Samuelsson, Antonio Ciarlo, Giuseppe Pesce, David Bronte Ciriza, Alessandro Magazzù, Onofrio M. Maragò, Antonio Sasso, Giovanni Volpe
Submitted to SPIE-ETAI
Date: 23 August 2022
Time: 13:40 (PDT)
Intracavity optical tweezers have been proven successful for trapping microscopic particles at very low average power intensity – much lower than the one in standard optical tweezers. This feature makes them particularly promising for the study of biological samples. The modeling of such systems, though, requires time-consuming numerical simulations that affect its usability and predictive power. With the help of machine learning, we can overcome the numerical bottleneck – the calculation of optical forces, torques, and losses – reproduce the results in the literature and generalize to the case of counterpropagating-beams intracavity optical trapping.
Presentation by H. Klein Moberg at SPIE-ETAI, San Diego, 23 August 2022
Henrik Klein Moberg, Christoph Langhammer, Daniel Midtvedt, Barbora Spackova, Bohdan Yeroshenko, David Albinsson, Joachim Fritzsche, Giovanni Volpe
Submitted to SPIE-ETAI
Date: 23 August 2022
Time: 9:15 (PDT)
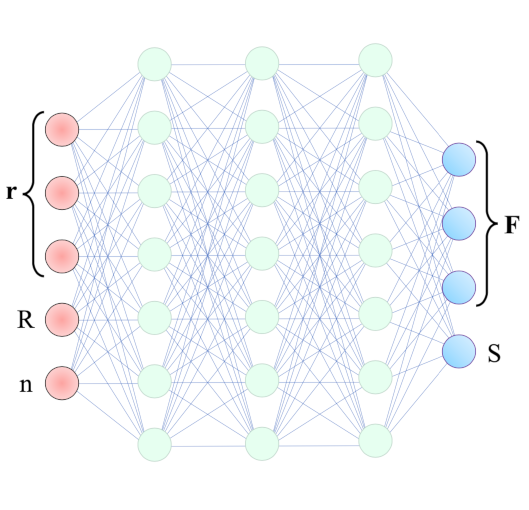
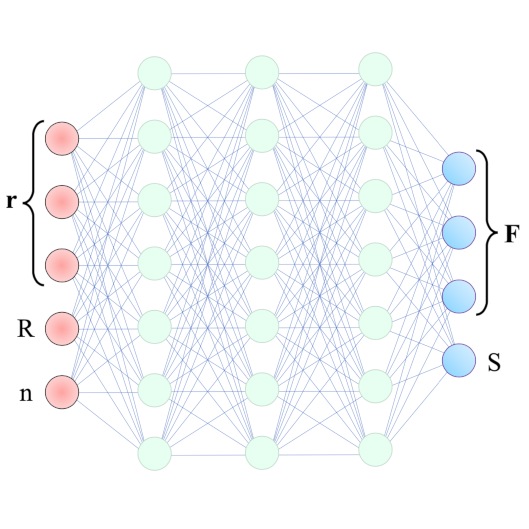
We show that a custom ResNet-inspired CNN architecture trained on simulated biomolecule trajectories surpasses the performance of standard algorithms in terms of tracking and determining the molecular weight and hydrodynamic radius of biomolecules in the low-kDa regime in NSM optical microscopy. We show that high accuracy and precision is retained even below the 10-kDa regime, constituting approximately an order of magnitude improvement in limit of detection compared to current state-of-the-art, enabling analysis of hitherto elusive species of biomolecules such as cytokines (~5-25 kDa) important for cancer research and the protein hormone insulin (~5.6 kDa), potentially opening up entirely new avenues of biological research.
Poster Presentation by F. Skärberg at SPIE-ETAI, San Diego, 22 August 2022
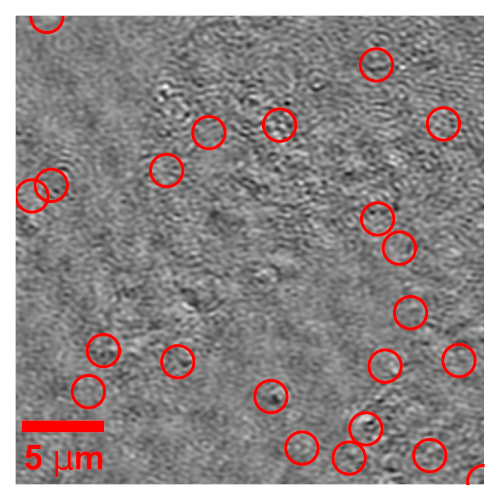
Fredrik Skärberg, Erik Olsén, Benjamin Midtvedt, Emelie V. Wesén, Elin K. Esbjörner, Giovanni Volpe, Fredrik Höök, Daniel Midtvedt
Submitted to SPIE-ETAI
Date: 22 August 2022
Time: 17:30 (PDT)
Optical characterization of nanoparticles in/outside cells is a challenging task, as the scattering from the nanoparticles is distorted by the scattering from the cells. In this work, we demonstrate a framework for optical mass quantification of intra- and extracellular nanoparticles by leveraging a novel deep learning method, LodeSTAR, in combination with off-axis twilight holography. The result provides new means for the exploration of nanoparticle/cell interactions.
Poster Presentation by Z. Korczak at SPIE-ETAI, San Diego, 22 August 2022
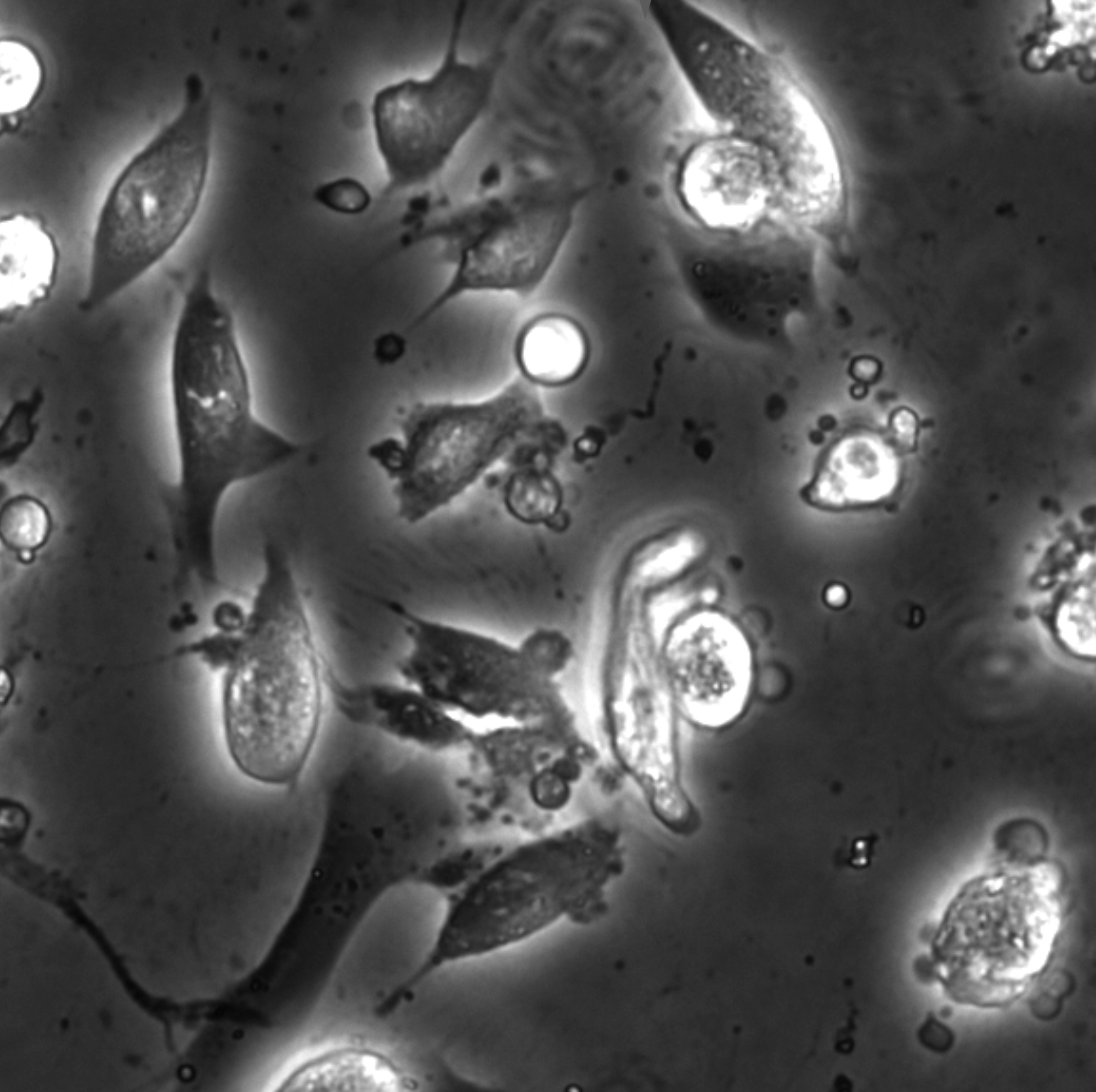
Zofia Korczak, Jesús D. Pineda, Saga Helgadottir, Benjamin Midtvedt, Mattias Goksör, Giovanni Volpe, Caroline B. Adiels
Submitted to SPIE-ETAI
Date: 22 August 2022
Time: 17:30 (PDT)
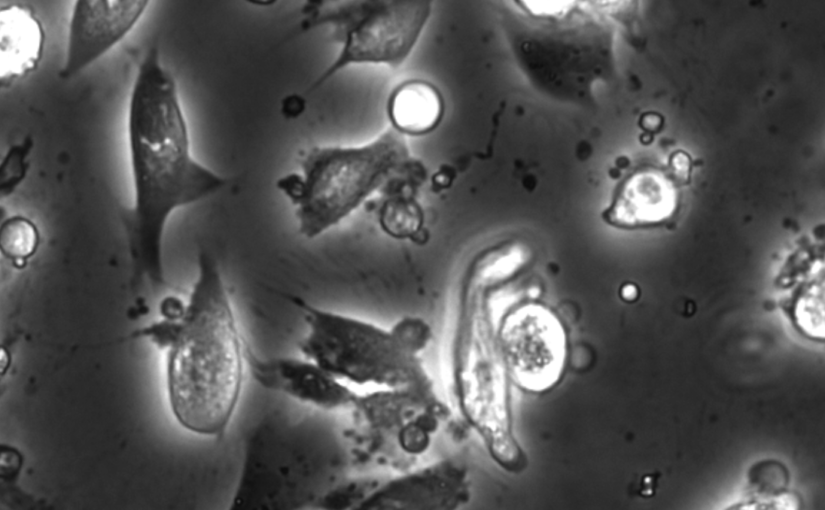
In vitro cell cultures rely on that the cultured cells thrive and behave in a physiologically relevant way. A standard approach to evaluate cells behaviour is to perform chemical staining in which fluorescent probes are added to the cell culture for further imaging and analysis. However, such a technique is invasive and sometimes even toxic to cells, hence, alternative methods are requested. Here, we describe an analysis method for the detection and discrimination of live and apoptotic cells using deep learning. This approach is less labour-intensive than traditional chemical staining procedures and enables cell imaging with minimal impact.
Soft Matter Lab members present at SPIE Optics+Photonics conference in San Diego, 21-25 August 2022
The Soft Matter Lab participates to the SPIE Optics+Photonics conference in San Diego, CA, USA, 21-25 August 2022, with the presentations listed below.
- Martin Selin: Scalable construction of quantum dot arrays using optical tweezers and deep learning (@SPIE)
22 August 2022 • 11:05 AM – 11:25 AM PDT | Conv. Ctr. Room 5A
- Fredrik Skärberg: Holographic characterisation of biological nanoparticles using deep learning (@SPIE)
22 August 2022 • 5:30 PM – 7:30 PM PDT | Conv. Ctr. Exhibit Hall B1
- Zofia Korczak: Dynamic virtual live/apoptotic cell assay using deep learning (@SPIE)
22 August 2022 • 5:30 PM – 7:30 PM PDT | Conv. Ctr. Exhibit Hall B1
- Henrik Klein Moberg: Seeing the invisible: deep learning optical microscopy for label-free biomolecule screening in the sub-10 kDa regime (@SPIE)
23 August 2022 • 9:15 AM – 9:35 AM PDT | Conv. Ctr. Room 5A
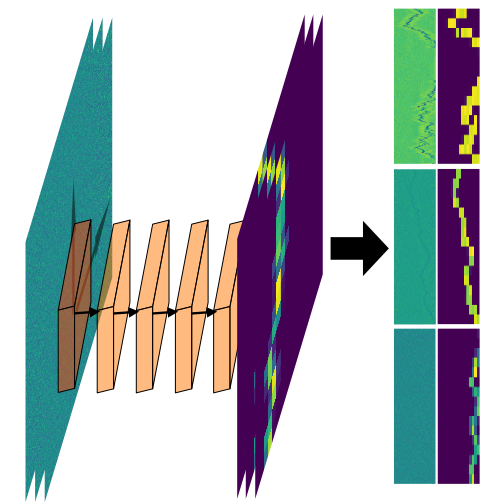
- Jesus Pineda: Revealing the spatiotemporal fingerprint of microscopic motion using geometric deep learning (@SPIE)
23 August 2022 • 11:05 AM – 11:25 AM PDT | Conv. Ctr. Room 5A
- Agnese Callegari: Simulating intracavity optical trapping with machine learning (@SPIE)
23 August 2022 • 1:40 PM – 2:00 PM PDT | Conv. Ctr. Room 5A
- Benjamin Midtvedt: Single-shot self-supervised particle tracking (@SPIE)
23 August 2022 • 2:20 PM – 2:40 PM PDT | Conv. Ctr. Room 5A
- Yu-Wei Chang: Neural network training with highly incomplete medical datasets (@SPIE)
24 August 2022 • 8:00 AM – 8:20 AM PDT | Conv. Ctr. Room 5A
- Daniel Midtvedt: Label-free characterization of biological matter across scales (@SPIE)
24 August 2022 • 9:10 AM – 9:40 AM PDT | Conv. Ctr. Room 5A
- Harshith Bachimanchi: Quantitative microplankton tracking by holographic microscopy and deep learning (@SPIE)
24 August 2022 • 11:45 AM – 12:05 PM PDT | Conv. Ctr. Room 5A
- Pablo Emiliano Gomez Ruiz: BRAPH 2.0: brain connectivity software with multilayer network analyses and machine learning (@SPIE)
24 August 2022 • 2:05 PM – 2:25 PM PDT | Conv. Ctr. Room 5A
- Yu-Wei Chang: Deep-learning analysis in tau PET for Alzheimer’s continuum (@SPIE)
24 August 2022 • 4:40 PM – 5:00 PM PDT | Conv. Ctr. Room 5A
Giovanni Volpe is also co-author of the presentations:
- Antonio Ciarlo: Periodic feedback effect in counterpropagating intracavity optical tweezers (@SPIE)
24 August 2022 • 2:00 PM – 2:20 PM PDT | Conv. Ctr. Room 1B
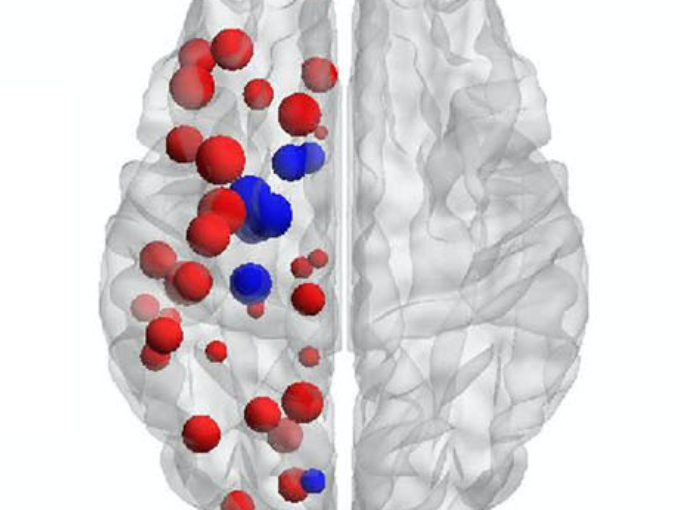
- Anna Canal Garcia: Multilayer brain connectivity analysis in Alzheimer’s disease using functional MRI data (@SPIE)
24 August 2022 • 2:25 PM – 2:45 PM PDT | Conv. Ctr. Room 5A
- Mite Mijalkov: A novel method for quantifying men and women-like features in brain structure and function (@SPIE)
24 August 2022 • 3:05 PM – 3:25 PM PDT | Conv. Ctr. Room 5A
- Carlo Manzo: An anomalous competition: assessment of methods for anomalous diffusion through a community effort (@SPIE)
25 August 2022 • 9:00 AM – 9:30 AM PDT | Conv. Ctr. Room 5A
- Stefano Bo: Characterization of anomalous diffusion with neural networks (@SPIE)
25 August 2022 • 10:00 AM – 10:30 AM PDT | Conv. Ctr. Room 5A
Invited Talk by G. Volpe at UCLA, 19 August 2022
Quantitative Digital Microscopy with Deep Learning
Giovanni Volpe
19 August 2022, 14:40 (PDT)
At the intersection of Photonics, Neuroscience, and AI
Ozcan Lab, UCLA, 19 August 2022
Video microscopy has a long history of providing insights and breakthroughs for a broad range of disciplines, from physics to biology. Image analysis to extract quantitative information from video microscopy data has traditionally relied on algorithmic approaches, which are often difficult to implement, time consuming, and computationally expensive. Recently, alternative data-driven approaches using deep learning have greatly improved quantitative digital microscopy, potentially offering automatized, accurate, and fast image analysis. However, the combination of deep learning and video microscopy remains underutilized primarily due to the steep learning curve involved in developing custom deep-learning solutions. To overcome this issue, we have introduced a software, currently at version DeepTrack 2.1, to design, train and validate deep-learning solutions for digital microscopy. We use it to exemplify how deep learning can be employed for a broad range of applications, from particle localization, tracking and characterization to cell counting and classification. Thanks to its user-friendly graphical interface, DeepTrack 2.1 can be easily customized for user-specific applications, and, thanks to its open-source object-oriented programming, it can be easily expanded to add features and functionalities, potentially introducing deep-learning-enhanced video microscopy to a far wider audience.
Multi-cohort and longitudinal Bayesian clustering study of stage and subtype in Alzheimer’s disease published in Nature Communications
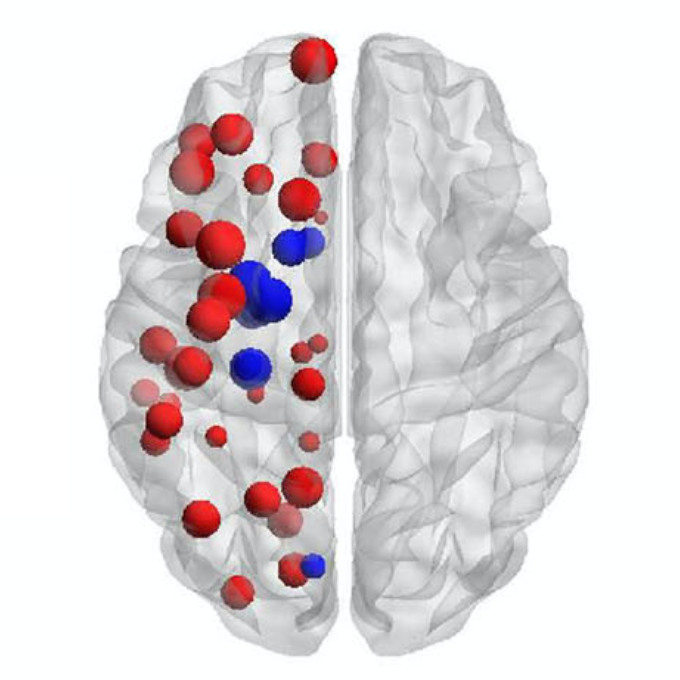
Konstantinos Poulakis, Joana B. Pereira, J.-Sebastian Muehlboeck, Lars-Olof Wahlund, Örjan Smedby, Giovanni Volpe, Colin L. Masters, David Ames, Yoshiki Niimi, Takeshi Iwatsubo, Daniel Ferreira, Eric Westman, Japanese Alzheimer’s Disease Neuroimaging Initiative & Australian Imaging, Biomarkers and Lifestyle study
Nature Communications 13, 4566 (2022)
doi: 10.1038/s41467-022-32202-6
Understanding Alzheimer’s disease (AD) heterogeneity is important for understanding the underlying pathophysiological mechanisms of AD. However, AD atrophy subtypes may reflect different disease stages or biologically distinct subtypes. Here we use longitudinal magnetic resonance imaging data (891 participants with AD dementia, 305 healthy control participants) from four international cohorts, and longitudinal clustering to estimate differential atrophy trajectories from the age of clinical disease onset. Our findings (in amyloid-β positive AD patients) show five distinct longitudinal patterns of atrophy with different demographical and cognitive characteristics. Some previously reported atrophy subtypes may reflect disease stages rather than distinct subtypes. The heterogeneity in atrophy rates and cognitive decline within the five longitudinal atrophy patterns, potentially expresses a complex combination of protective/risk factors and concomitant non-AD pathologies. By alternating between the cross-sectional and longitudinal understanding of AD subtypes these analyses may allow better understanding of disease heterogeneity.