Daniel Andrén, Denis G. Baranov, Steven Jones, Giovanni Volpe, Ruggero Verre, Mikael Käll
Nat. Nanotechnol. (2021)
doi: 10.1038/s41565-021-00941-0
arXiv: 2012.10205
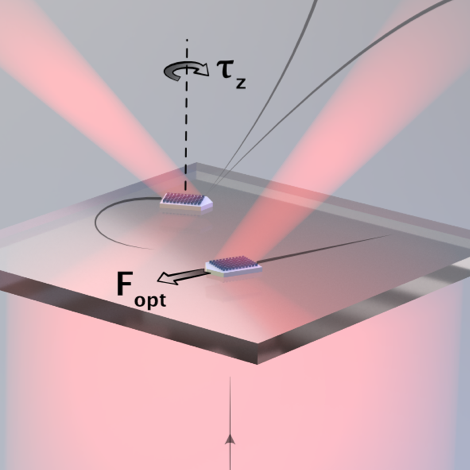
Nanostructured dielectric metasurfaces offer unprecedented opportunities to manipulate light by imprinting an arbitrary phase gradient on an impinging wavefront. This has resulted in the realization of a range of flat analogues to classical optical components, such as lenses, waveplates and axicons. However, the change in linear and angular optical momentum associated with phase manipulation also results in previously unexploited forces and torques that act on the metasurface itself. Here we show that these optomechanical effects can be utilized to construct optical metavehicles – microscopic particles that can travel long distances under low-intensity plane-wave illumination while being steered by the polarization of the incident light. We demonstrate movement in complex patterns, self-correcting motion and an application as transport vehicles for microscopic cargoes, which include unicellular organisms. The abundance of possible optical metasurfaces attests to the prospect of developing a wide variety of metavehicles with specialized functional behaviours.