Seven members of the Soft Matter Lab (Saga Helgadottir, Benjamin Midtvedt, Aykut Argun, Laura Pérez-Garcia, Daniel Midtvedt, Harshith Bachimanchi, Emiliano Gómez) were selected for oral and poster presentations at the SPIE Optics+Photonics Digital Forum, August 24-28, 2020.
The SPIE digital forum is a free, online only event.
The registration for the Digital Forum includes access to all presentations and proceedings.
The Soft Matter Lab contributions are part of the SPIE Nanoscience + Engineering conferences, namely the conference on Emerging Topics in Artificial Intelligence 2020 and the conference on Optical Trapping and Optical Micromanipulation XVII.
The contributions being presented are listed below, including also the presentations co-authored by Giovanni Volpe.
Note: the presentation times are indicated according to PDT (Pacific Daylight Time) (GMT-7)
Saga Helgadottir
Digital video microscopy with deep learning (Invited Paper)
26 August 2020, 10:30 AM
SPIE Link: here.
Aykut Argun
Calibration of force fields using recurrent neural networks
26 August 2020, 8:30 AM
SPIE Link: here.
Laura Pérez-García
Deep-learning enhanced light-sheet microscopy
25 August 2020, 9:10 AM
SPIE Link: here.
Daniel Midtvedt
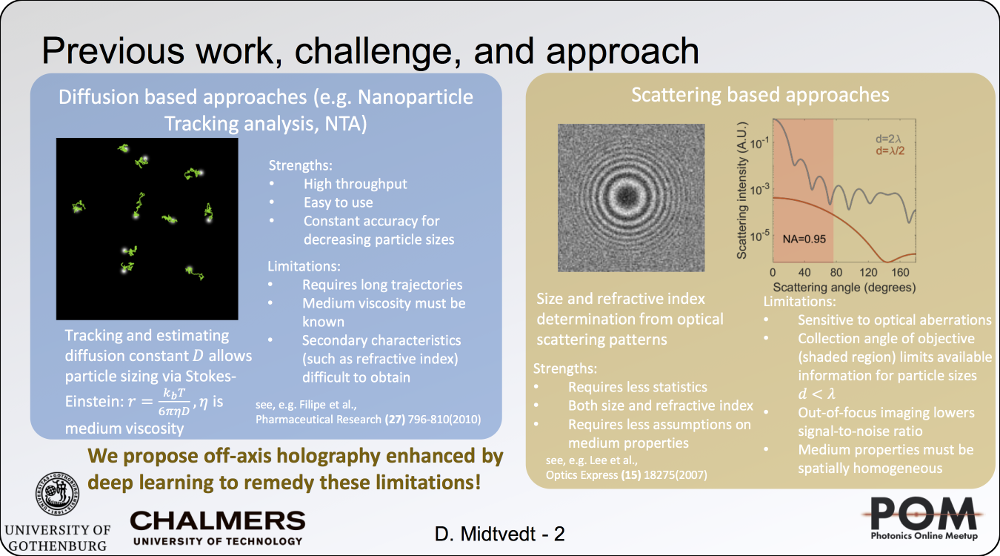
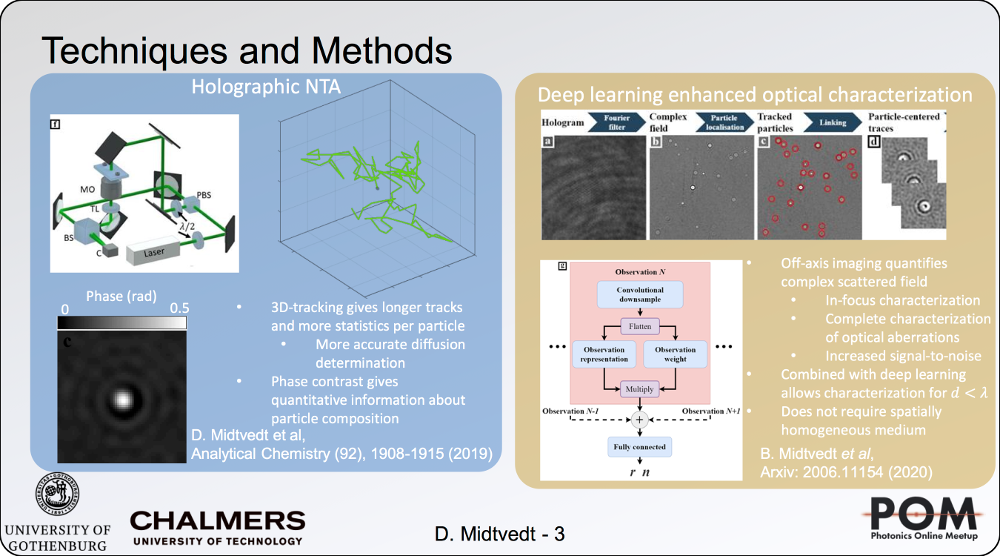
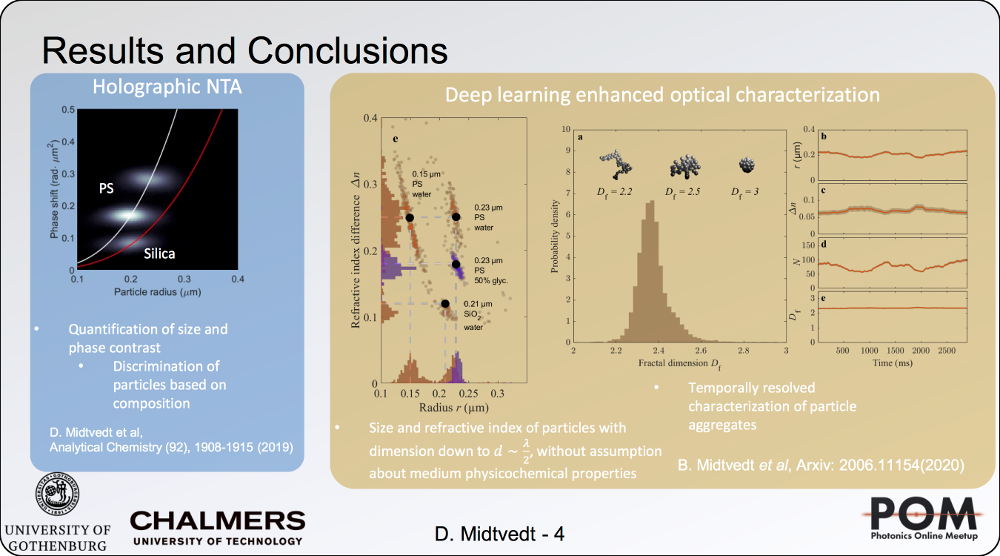
Holographic characterization of subwavelength particles enhanced by deep learning
24 August 2020, 2:40 PM
SPIE Link: here.
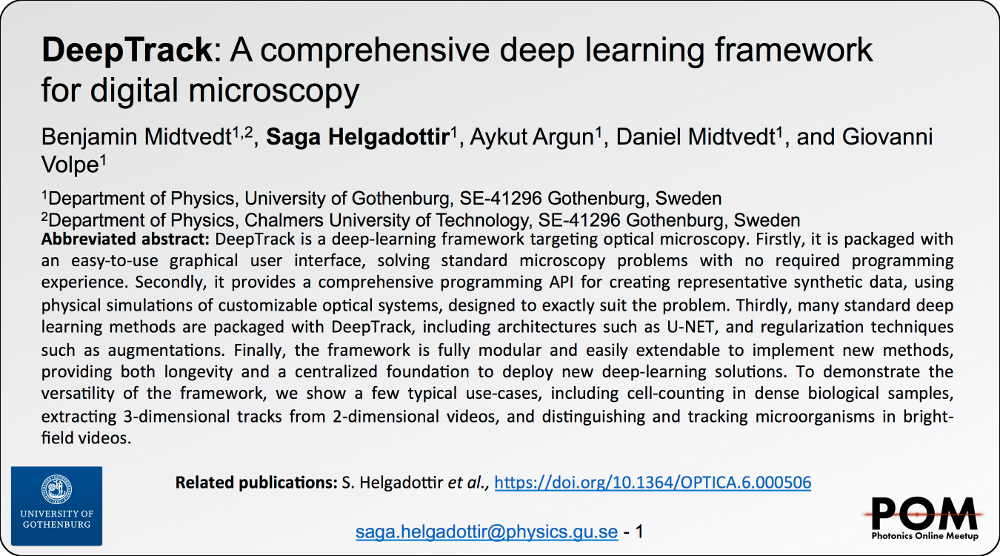
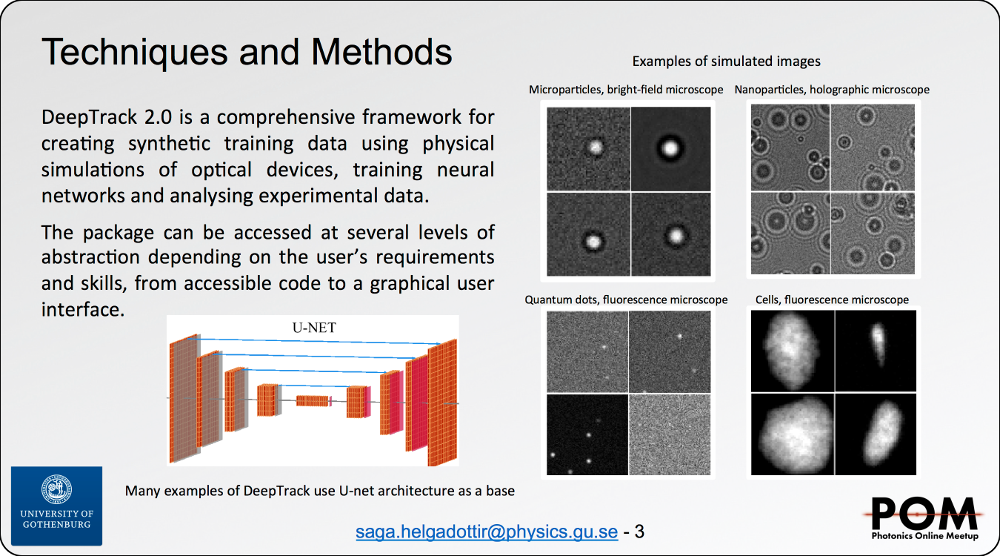
Benjamin Midtvedt
DeepTrack: A comprehensive deep learning framework for digital microscopy
26 August 2020, 11:40 AM
SPIE Link: here.
Gorka Muñoz-Gil
The anomalous diffusion challenge: Single trajectory characterisation as a competition
26 August 2020, 12:00 PM
SPIE Link: here.
Meera Srikrishna
Brain tissue segmentation using U-Nets in cranial CT scans
25 August 2020, 2:00 PM
SPIE Link: here.
Juan S. Sierra
Automated corneal endothelium image segmentation in the presence of cornea guttata via convolutional neural networks
26 August 2020, 11:50 AM
SPIE Link: here.
Harshith Bachimanchi
Digital holographic microscopy driven by deep learning: A study on marine planktons (Poster)
24 August 2020, 5:30 PM
SPIE Link: here.
Emiliano Gómez
BRAPH 2.0: Software for the analysis of brain connectivity with graph theory (Poster)
24 August 2020, 5:30 PM
SPIE Link: here.
Laura Pérez-García
Reconstructing complex force fields with optical tweezers
24 August 2020, 5:00 PM
SPIE Link: here.
Alejandro V. Arzola
Direct visualization of the spin-orbit angular momentum conversion in optical trapping
25 August 2020, 10:40 AM
SPIE Link: here.
Isaac Lenton
Illuminating the complex behaviour of particles in optical traps with machine learning
26 August 2020, 9:10 AM
SPIE Link: here.
Fatemeh Kalantarifard
Optical trapping of microparticles and yeast cells at ultra-low intensity by intracavity nonlinear feedback forces
24 August 2020, 11:10 AM
SPIE Link: here.
Note: the presentation times are indicated according to PDT (Pacific Daylight Time) (GMT-7)
 Gustaf Sjösten joined the Soft Matter Lab on 1 September 2020.
Gustaf Sjösten joined the Soft Matter Lab on 1 September 2020.