Intercellular communication induces glycolytic synchronization waves between individually oscillating cells
Martin Mojica-Benavides, David D. van Niekerk, Mite Mijalkov, Jacky L. Snoep, Bernhard Mehlig, Giovanni Volpe, Caroline B. Adiels & Mattias Goksör
PNAS 118(6), e2010075118 (2021)
doi: 10.1073/pnas.2010075118
arXiv: 1909.05187
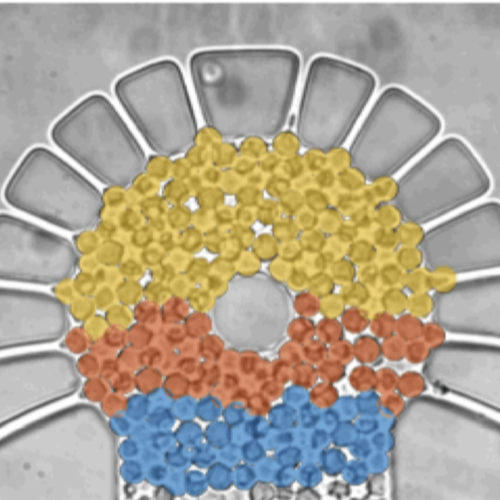
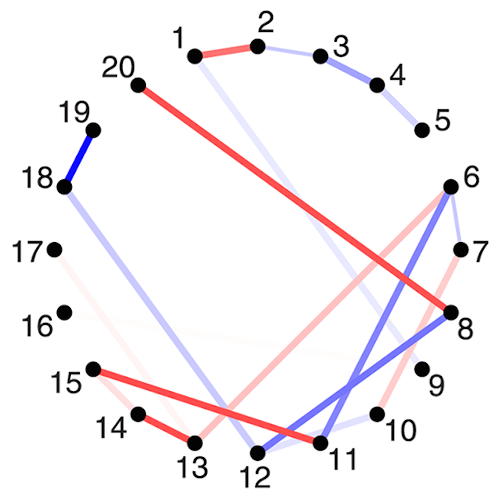
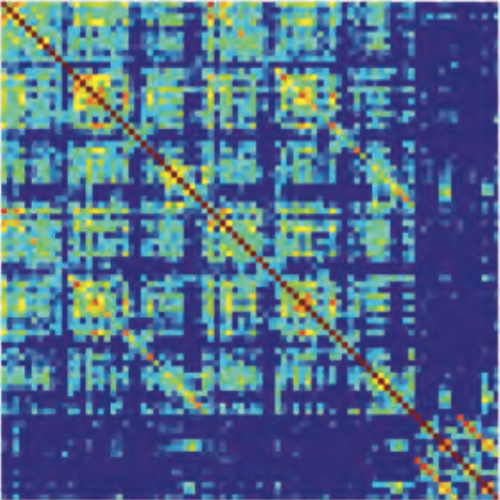
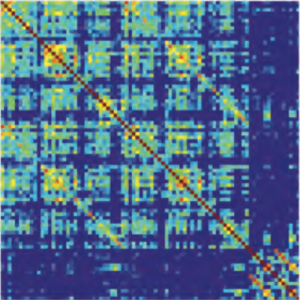
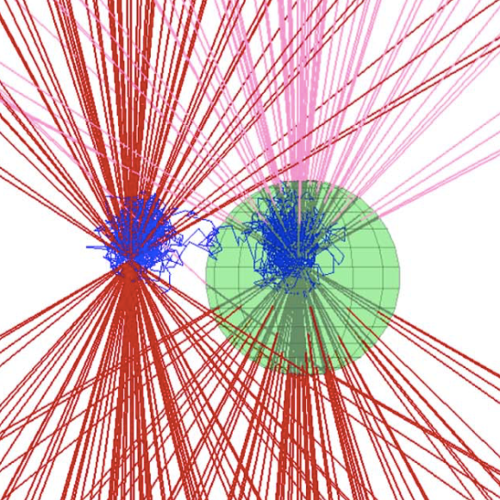
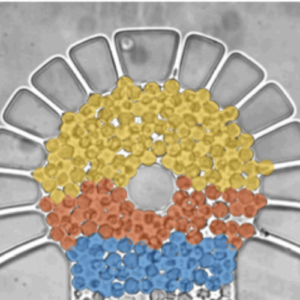
Metabolic oscillations in single cells underlie the mechanisms behind cell synchronization and cell-cell communication. For example, glycolytic oscillations mediated by biochemical communication between cells may synchronize the pulsatile insulin secretion by pancreatic tissue, and a link between glycolytic synchronization anomalies and type-2 diabetes has been hypotesized. Cultures of yeast cells have provided an ideal model system to study synchronization and propagation waves of glycolytic oscillations in large populations. However, the mechanism by which synchronization occurs at individual cell-cell level and overcome local chemical concentrations and heterogenic biological clocks, is still an open question because of experimental limitations in sensitive and specific handling of single cells. Here, we show how the coupling of intercellular diffusion with the phase regulation of individual oscillating cells induce glycolytic synchronization waves. We directly measure the single-cell metabolic responses from yeast cells in a microfluidic environment and characterize a discretized cell-cell communication using graph theory. We corroborate our findings with simulations based on a kinetic detailed model for individual yeast cells. These findings can provide insight into the roles cellular synchronization play in biomedical applications, such as insulin secretion regulation at the cellular level.