Mite Mijalkov, Giovanni Volpe, Joana B Pereira
Anna Canal-Garcia, Emiliano Gómez-Ruiz, Mite Mijalkov, Yu-Wei Chang, Giovanni Volpe, Joana B Pereira, Alzheimer’s Disease Neuroimaging Initiative
Cerebral Cortex, bhab429 (2022)
doi: 10.1093/cercor/bhab429
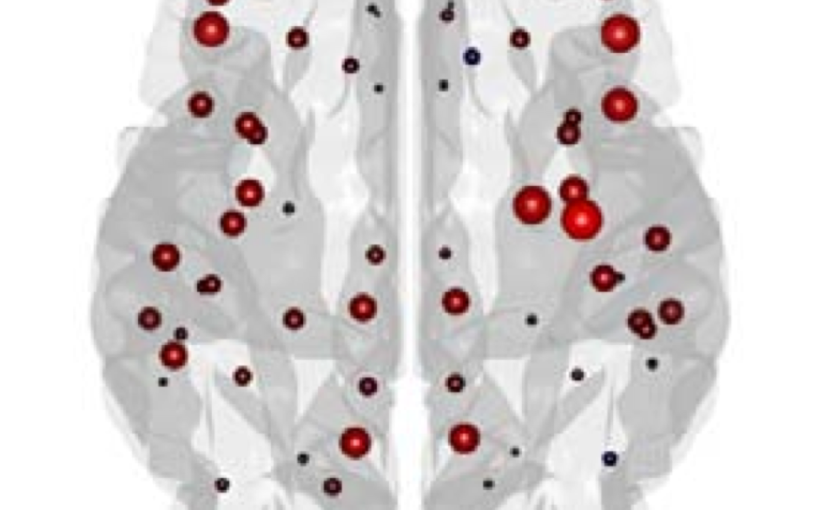
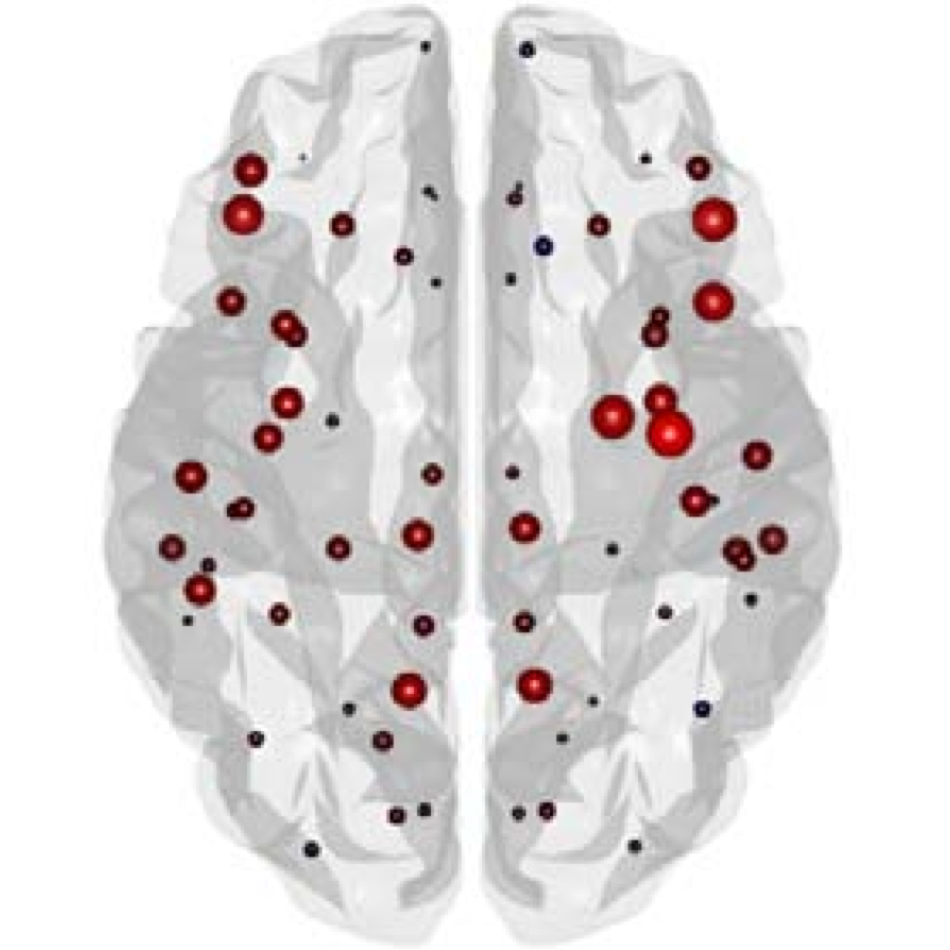
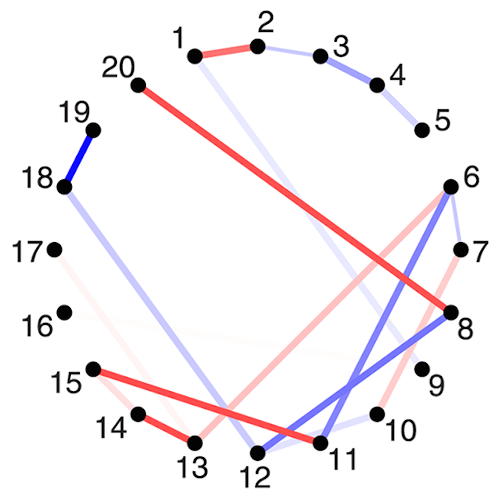
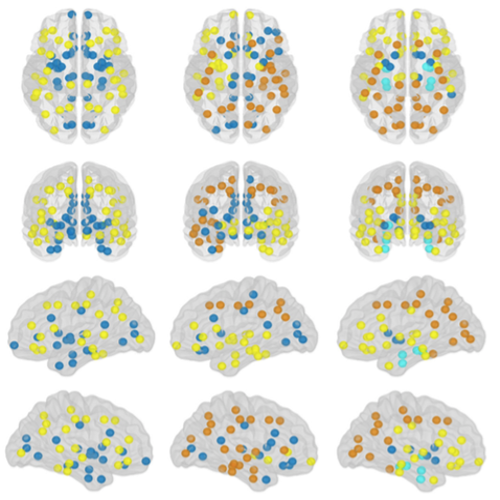
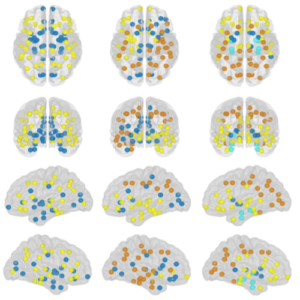
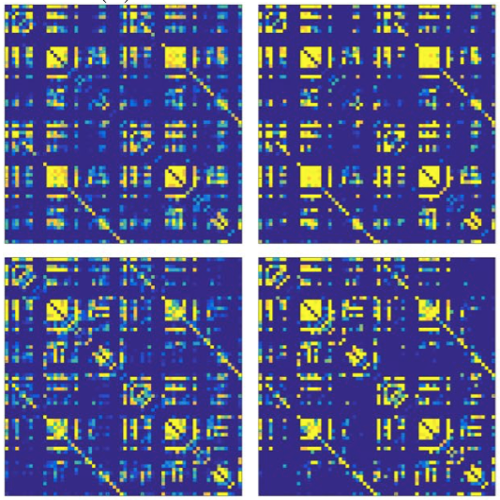
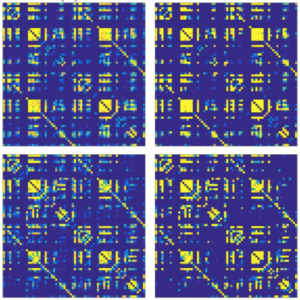
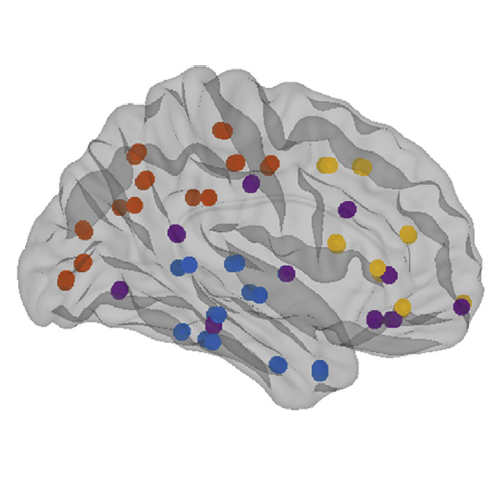
The organization of the Alzheimer’s disease (AD) connectome has been studied using graph theory using single neuroimaging modalities such as positron emission tomography (PET) or structural magnetic resonance imaging (MRI). Although these modalities measure distinct pathological processes that occur in different stages in AD, there is evidence that they are not independent from each other. Therefore, to capture their interaction, in this study we integrated amyloid PET and gray matter MRI data into a multiplex connectome and assessed the changes across different AD stages. We included 135 cognitively normal (CN) individuals without amyloid-β pathology (Aβ−) in addition to 67 CN, 179 patients with mild cognitive impairment (MCI) and 132 patients with AD dementia who all had Aβ pathology (Aβ+) from the Alzheimer’s Disease Neuroimaging Initiative. We found widespread changes in the overlapping connectivity strength and the overlapping connections across Aβ-positive groups. Moreover, there was a reorganization of the multiplex communities in MCI Aβ + patients and changes in multiplex brain hubs in both MCI Aβ + and AD Aβ + groups. These findings offer a new insight into the interplay between amyloid-β pathology and brain atrophy over the course of AD that moves beyond traditional graph theory analyses based on single brain networks.