Amyloid network topology characterizes the progression of Alzheimer’s disease during the predementia stages
Joana B. Pereira, Tor Olof Strandberg, Sebastian Palmqvist, Giovanni Volpe, Danielle van Westen, Eric Westman & Oskar Hansson, for the Alzheimer’s Disease Neuroimaging Initiative
Cerebral Cortex 28(1), 340—349 (2018)
DOI: 10.1093/cercor/bhx294
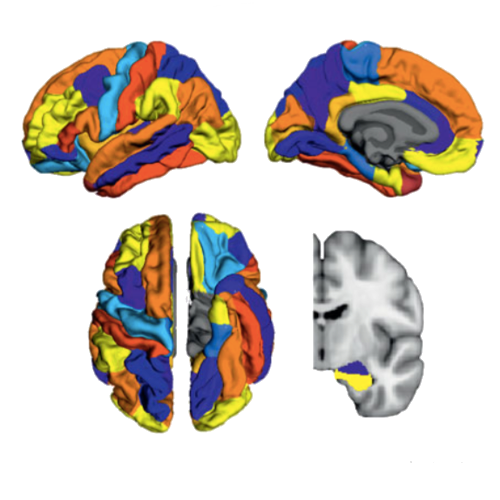
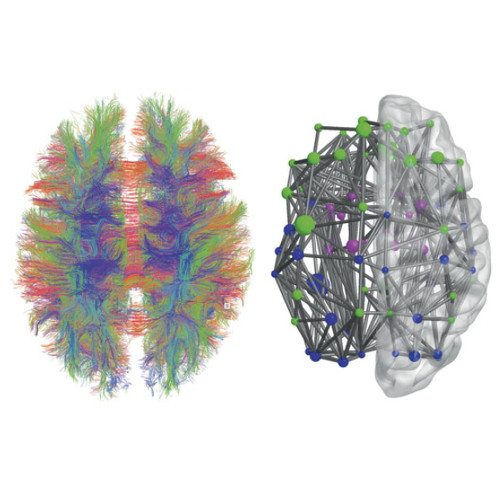
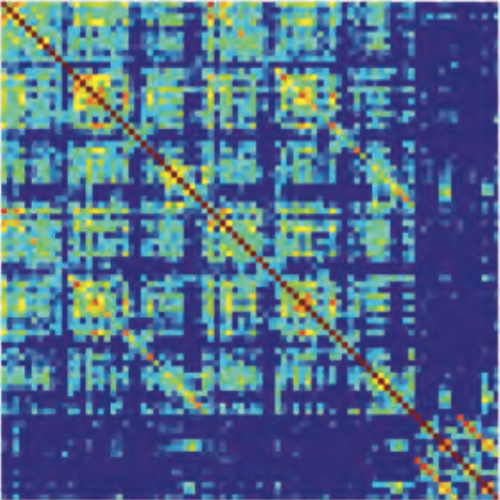
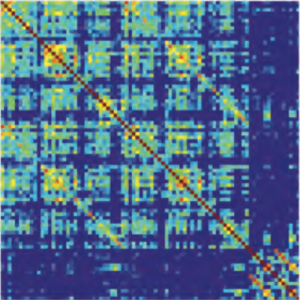
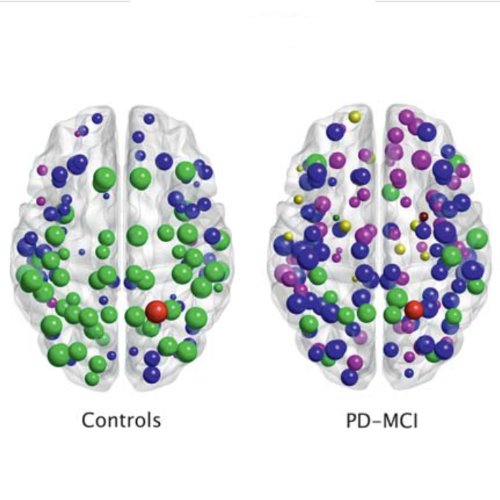
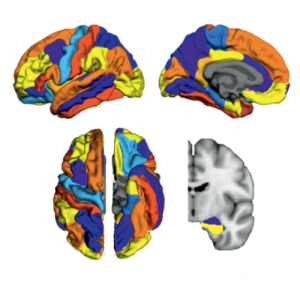
There is increasing evidence showing that the accumulation of the amyloid-β (Aβ) peptide into extracellular plaques is a central event in Alzheimer’s disease (AD). These abnormalities can be detected as lowered levels of Aβ42 in the cerebrospinal fluid (CSF) and are followed by increased amyloid burden on positron emission tomography (PET) several years before the onset of dementia. The aim of this study was to assess amyloid network topology in nondemented individuals with early stage Aβ accumulation, defined as abnormal CSF Aβ42 levels and normal Florbetapir PET (CSF+/PET−), and more advanced Aβ accumulation, defined as both abnormal CSF Aβ42 and Florbetapir PET (CSF+/PET+). The amyloid networks were built using correlations in the mean 18F-florbetapir PET values between 72 brain regions and analyzed using graph theory analyses. Our findings showed an association between early amyloid stages and increased covariance as well as shorter paths between several brain areas that overlapped with the default-mode network (DMN). Moreover, we found that individuals with more advanced amyloid accumulation showed more widespread changes in brain regions both within and outside the DMN. These findings suggest that amyloid network topology could potentially be used to assess disease progression in the predementia stages of AD.