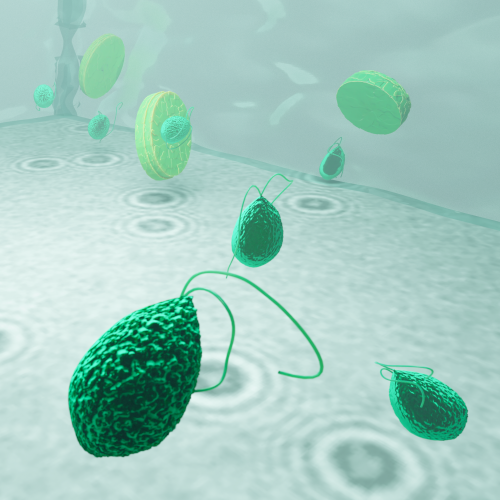
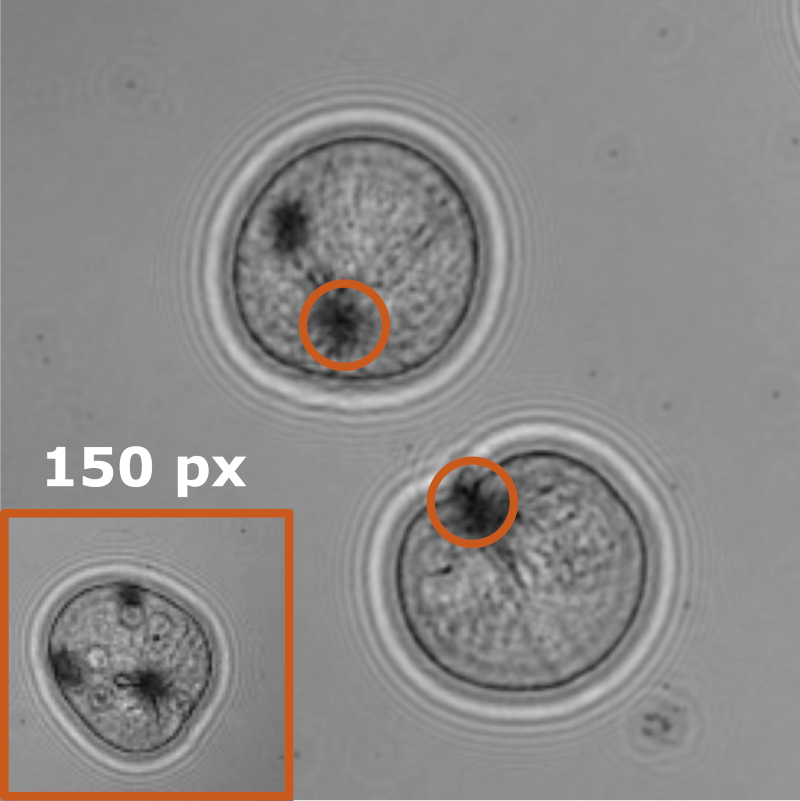
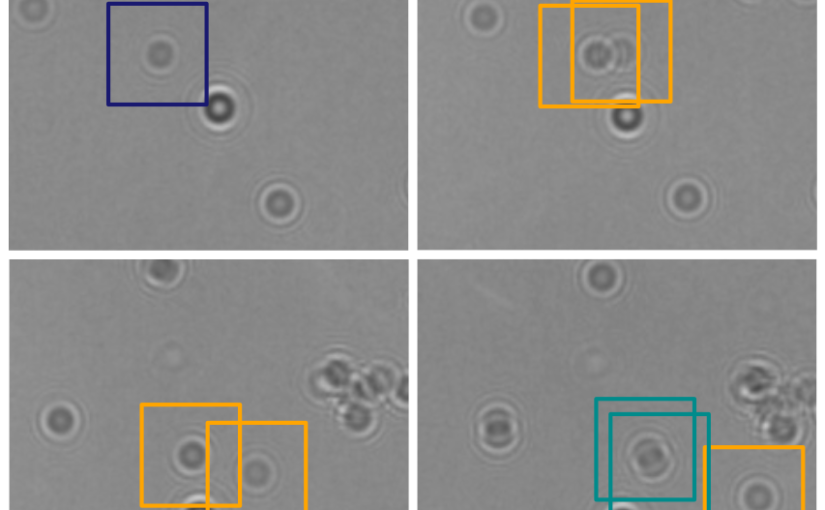
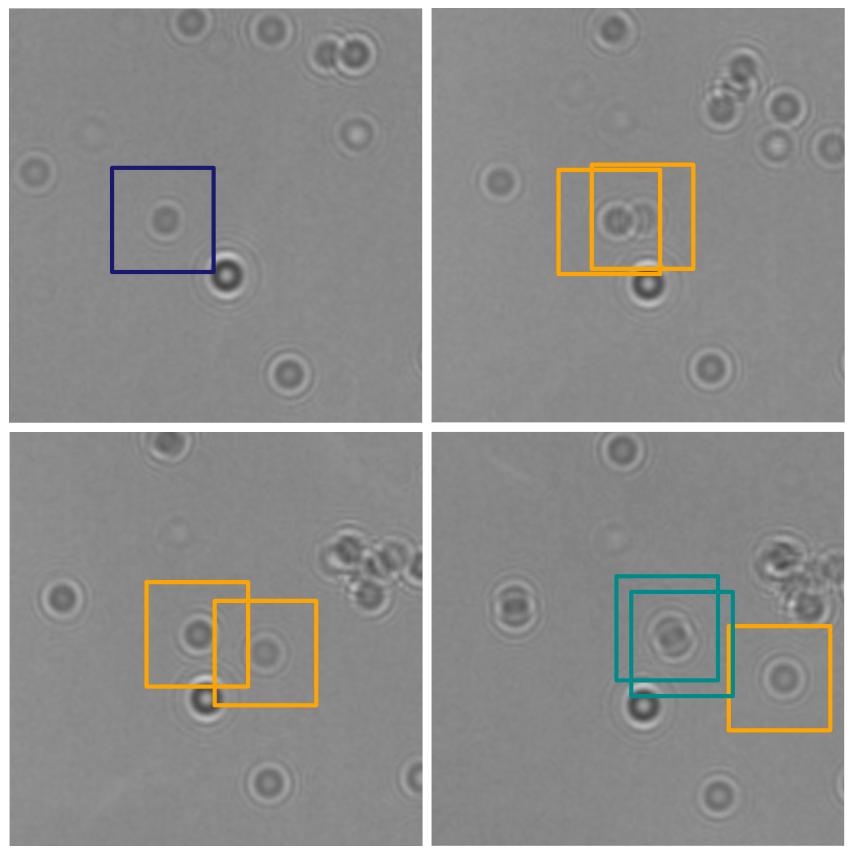
In this work, Harshith presented his recent work on combining holographic microscopy and deep learning to study the marine microplankton. He demonstrated that the combination of holographic microscopy and deep learning can be used to follow the marine microorganisms throughout their lifespan, continuously measuring their three-dimensional positions and dry mass. The deep-learning algorithms circumvent the computationally intensive processing of holographic data and allow rapid measurements over extended periods of time. This enables to reliably estimate growth rates, both in terms of dry mass increase and cell divisions, as well as to measure trophic interactions between species such as predation events. Studying individual interactions in idealized small systems provides insights that help us understand microbial food webs and ultimately larger-scale processes. He exemplified this by showing detailed descriptions of micro-zooplankton feeding events, cell divisions, and long-term monitoring of single cells from division to division.
The article related to this presentation can be found at the following link: Microplankton life histories revealed by holographic microscopy and deep learning.