Gan Wang, Marcel Rey, Antonio Ciarlo, Mohanmmad Mahdi Shanei, Kunli Xiong, Giuseppe Pesce, Mikael Käll and Giovanni Volpe
Date: 5 September 2024
Time: 15:45-16:00
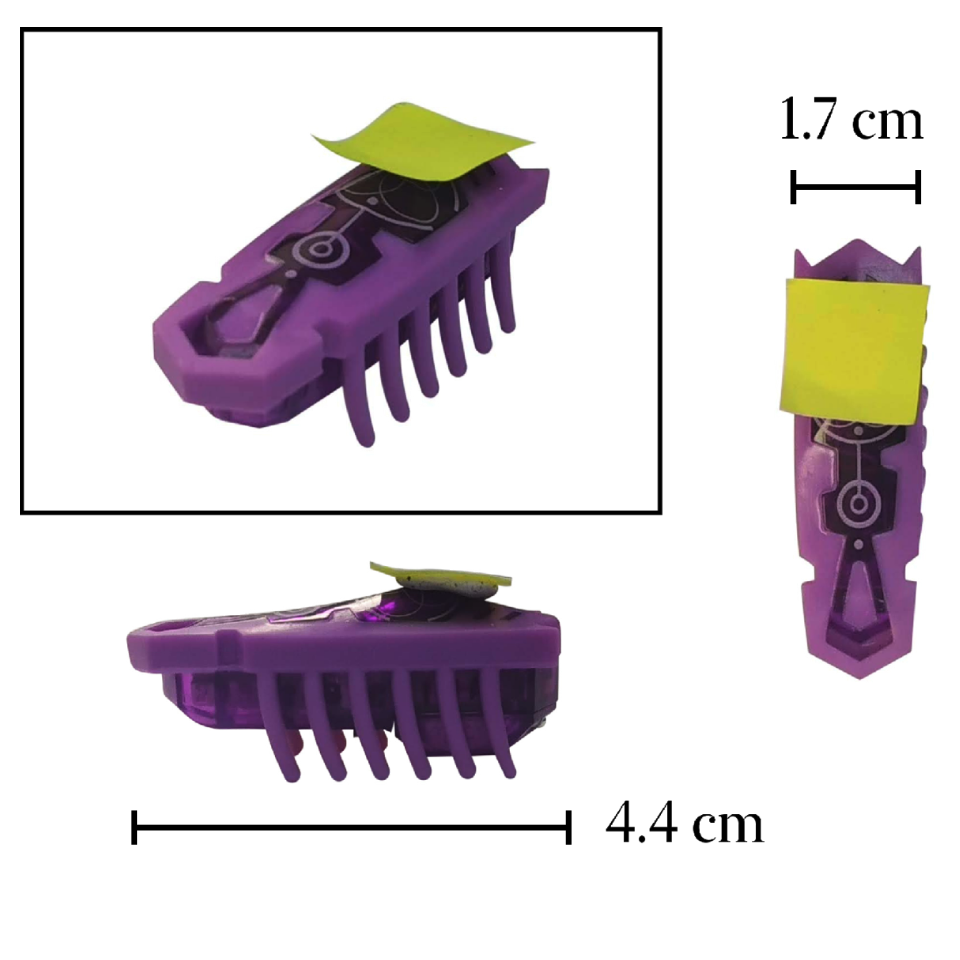
The incorporation of Moore’s law into integrated circuits has spurred opportunities for downsizing traditional mechanical components. Despite advancements in single on-chip motors using electrical, optical, and magnetic drives under ~100 μm, creating complex machines with multiple units remains challenging. Here, we developed a ~10 μm on-chip micromotor using a method compatible with complementary metal oxide semiconductors (CMOS) process. The meta-surface is embedded with the motor to control the incident laser beam direction, enabling momentum exchange with light for movement. The rotation direction and speed are adjustable through the meta-surface, along with the intensity and polarization of applied light. By combining these motors and controlling the configuration, we create complex machines with a size similar to traditional machines below 50um, such as the rotary motion mode of multiple gear coupled gear trains, and the linear motion mode combined with rack and pinion, and combine the micro metal The mirror is introduced into the machine to realize the self-controlled scanning function of the laser in a fixed area.