Saga Helgadottir, Benjamin Midtvedt, Jesús Pineda, Alan Sabirsh, Caroline B. Adiels, Stefano Romeo, Daniel Midtvedt, Giovanni Volpe
Biophysics Rev. 2, 031401 (2021)
arXiv: 2012.12986
doi: 10.1063/5.0044782
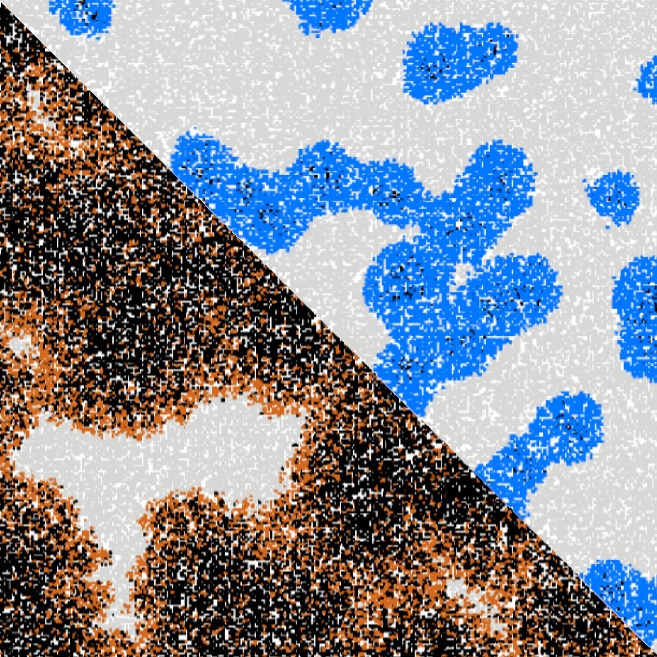
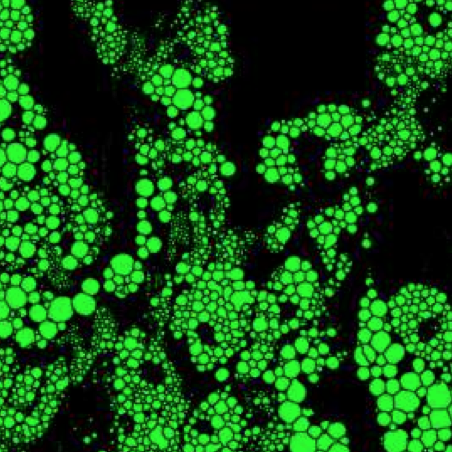
Quantitative analysis of cell structures is essential for biomedical and pharmaceutical research. The standard imaging approach relies on fluorescence microscopy, where cell structures of interest are labeled by chemical staining techniques. However, these techniques are often invasive and sometimes even toxic to the cells, in addition to being time-consuming, labor-intensive, and expensive. Here, we introduce an alternative deep-learning-powered approach based on the analysis of bright-field images by a conditional generative adversarial neural network (cGAN). We show that this approach can extract information from the bright-field images to generate virtually-stained images, which can be used in subsequent downstream quantitative analyses of cell structures. Specifically, we train a cGAN to virtually stain lipid droplets, cytoplasm, and nuclei using bright-field images of human stem-cell-derived fat cells (adipocytes), which are of particular interest for nanomedicine and vaccine development. Subsequently, we use these virtually-stained images to extract quantitative measures about these cell structures. Generating virtually-stained fluorescence images is less invasive, less expensive, and more reproducible than standard chemical staining; furthermore, it frees up the fluorescence microscopy channels for other analytical probes, thus increasing the amount of information that can be extracted from each cell.