Meera Srikrishna, Nicholas J. Ashton, Alexis Moscoso, Joana B. Pereira, Rolf A. Heckemann, Danielle van Westen, Giovanni Volpe, Joel Simrén, Anna Zettergren, Silke Kern, Lars-Olof Wahlund, Bibek Gyanwali, Saima Hilal, Joyce Chong Ruifen, Henrik Zetterberg, Kaj Blennow, Eric Westman, Christopher Chen, Ingmar Skoog, Michael Schöll
Alzheimer’s & Dementia 20, 629–640 (2024)
arXiv: 2401.06260
doi: 10.1002/alz.13445
INTRODUCTION
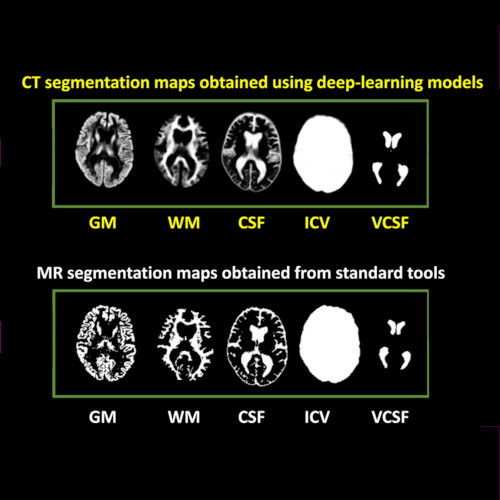
Cranial computed tomography (CT) is an affordable and widely available imaging modality that is used to assess structural abnormalities, but not to quantify neurodegeneration. Previously we developed a deep-learning–based model that produced accurate and robust cranial CT tissue classification.
MATERIALS AND METHODS
We analyzed 917 CT and 744 magnetic resonance (MR) scans from the Gothenburg H70 Birth Cohort, and 204 CT and 241 MR scans from participants of the Memory Clinic Cohort, Singapore. We tested associations between six CT-based volumetric measures (CTVMs) and existing clinical diagnoses, fluid and imaging biomarkers, and measures of cognition.
RESULTS
CTVMs differentiated cognitively healthy individuals from dementia and prodromal dementia patients with high accuracy levels comparable to MR-based measures. CTVMs were significantly associated with measures of cognition and biochemical markers of neurodegeneration.
DISCUSSION
These findings suggest the potential future use of CT-based volumetric measures as an informative first-line examination tool for neurodegenerative disease diagnostics after further validation.