Caroline B. Adiels
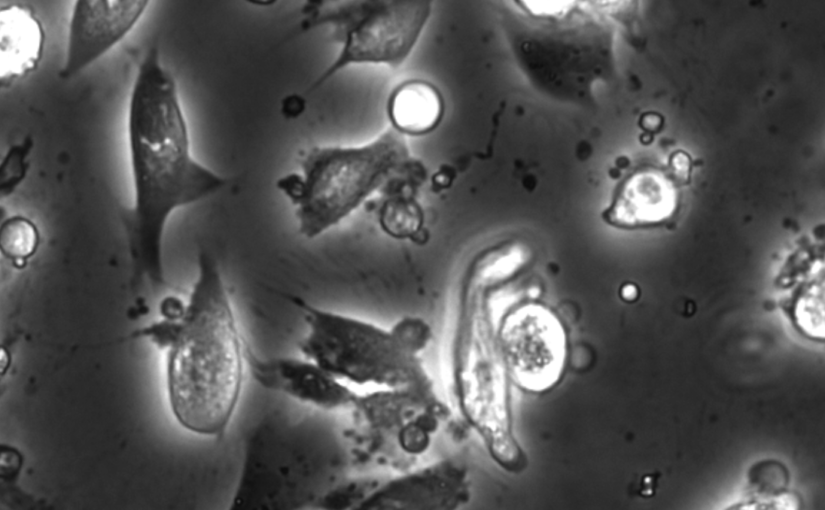
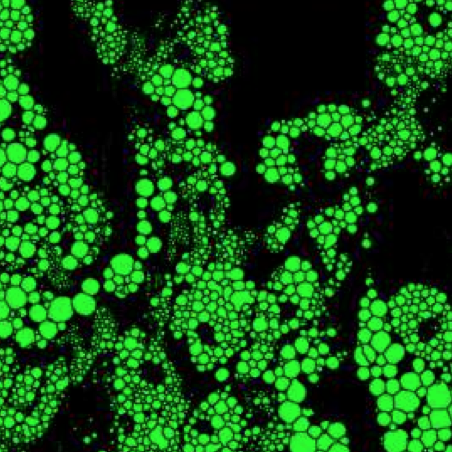
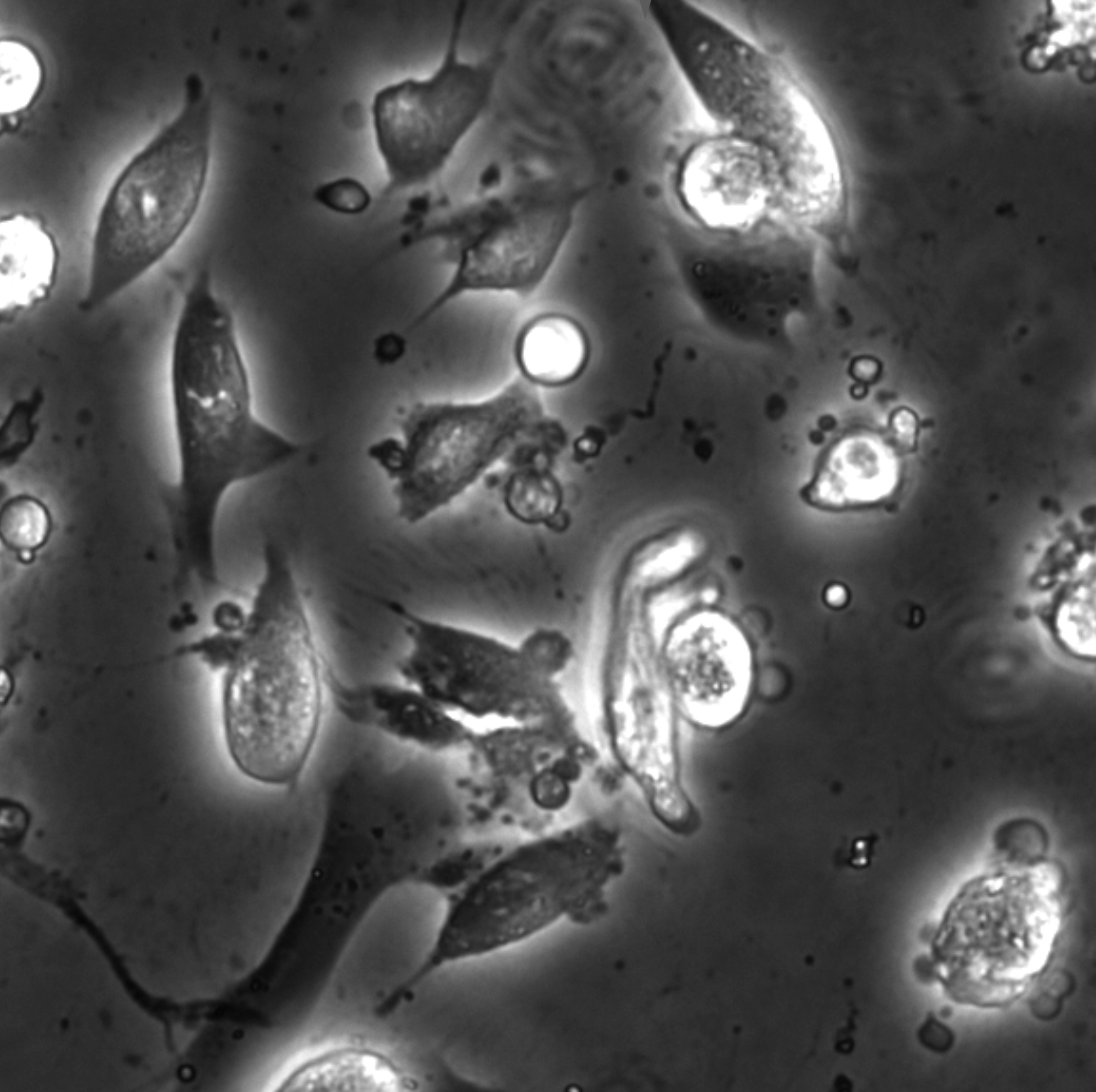
Chemical live/dead assay has a long history of providing information about the viability of cells cultured in vitro. The standard methods rely on imaging chemically-stained cells using fluorescence microscopy and further analysis of the obtained images to retrieve the proportion of living cells in the sample. However, such a technique is not only time-consuming but also invasive. Due to the toxicity of chemical dyes, once a sample is stained, it is discarded, meaning that longitudinal studies are impossible using this approach. Further, information about when cells start programmed cell death (apoptosis) is more relevant for dynamic studies. Here, we present an alternative method where cell images from phase-contrast time-lapse microscopy are virtually-stained using deep learning. In this study, human endothelial cells are stained live or apoptotic and subsequently counted using the self-supervised single-shot deep-learning technique (LodeSTAR). Our approach is less labour-intensive than traditional chemical staining procedures and provides dynamic live/apoptotic cell ratios from a continuous cell population with minimal impact. Further, it can be used to extract data from dense cell samples, where manual counting is unfeasible.
Date: 31 May 2023
Time: 10:30
Place: MC2 Kollektorn
Event: AI for Scientific Data Analysis: Miniconference