Ordering of Binary Colloidal Crystals by Random Potentials
André S. Nunes, Sabareesh K. P. Velu, Iryna Kasianiuk, Denys Kasyanyuk, Agnese Callegari, Giorgio Volpe, Margarida M. Telo da Gama, Giovanni Volpe & Nuno A. M. Araújo
Soft Matter 16, 4267-4273 (2020)
doi: https://doi.org/10.1039/D0SM00208A
arXiv: 1903.01579
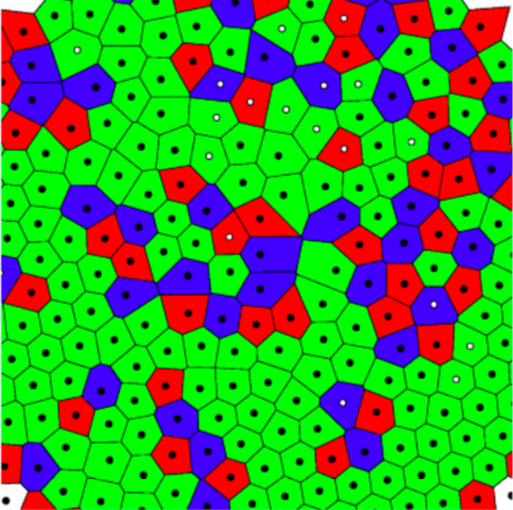
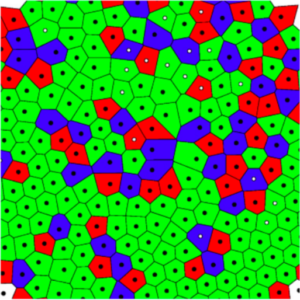
Structural defects are ubiquitous in condensed matter, and not always a nuisance. For example, they underlie phenomena such as Anderson localization and hyperuniformity, and they are now being exploited to engineer novel materials. Here, we show experimentally that the density of structural defects in a 2D binary colloidal crystal can be engineered with a random potential. We generate the random potential using an optical speckle pattern, whose induced forces act strongly on one species of particles (strong particles) and weakly on the other (weak particles). Thus, the strong particles are more attracted to the randomly distributed local minima of the optical potential, leaving a trail of defects in the crystalline structure of the colloidal crystal. While, as expected, the crystalline ordering initially decreases with an increasing fraction of strong particles, the crystalline order is surprisingly recovered for sufficiently large fractions. We confirm our experimental results with particle-based simulations, which permit us to elucidate how this non-monotonic behavior results from the competition between the particle-potential and particle-particle interactions.