Harshith Bachimanchi, Benjamin Midtvedt, Daniel Midtvedt, Erik Selander, and Giovanni Volpe
Presentation at ISMC 2022
Poznan, Poland
19 September 2022, 12:40 CEST
A droplet of sea water contains an entire ecosytem. There are microscopic plants, the phytoplanktons, which produce oxygen by absorbing carbon dioxide from the atmsphere by the process of photosynthesis. There are microscopic animals, the microzooplankton, which feed on the phytoplankton. In oceanic ecology, phytoplanktons consume around 65 peta grams of carbon annually, producing approximately 50% of oxygen on the Earth. Microzooplankton take on the role of herbivores, and consume about two thirds (40 Pg carbon) of this primary production. Despite their central importance, our understanding of the phytoplankton and microzooplankton in shaping oceanic communities is still much less developed at a single plankton level.
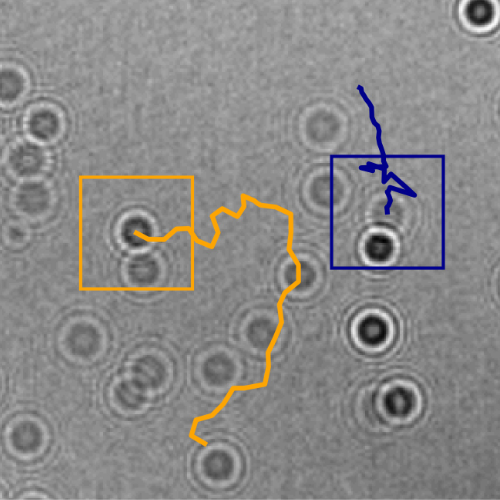
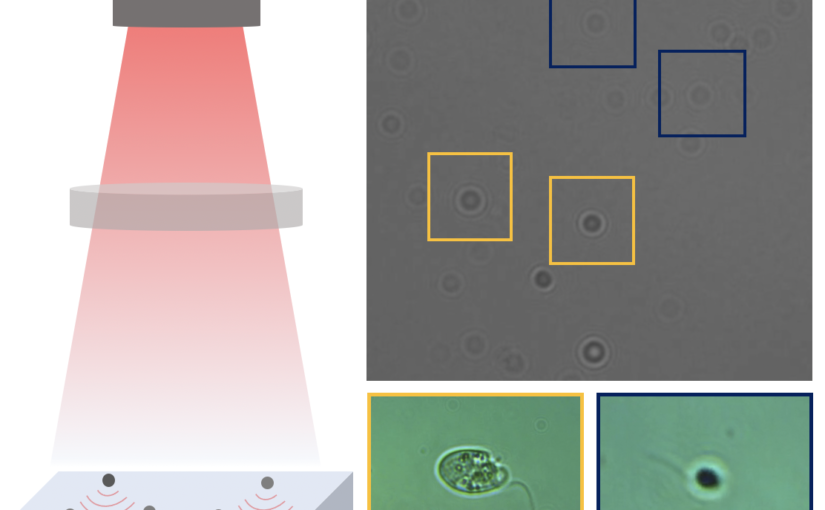
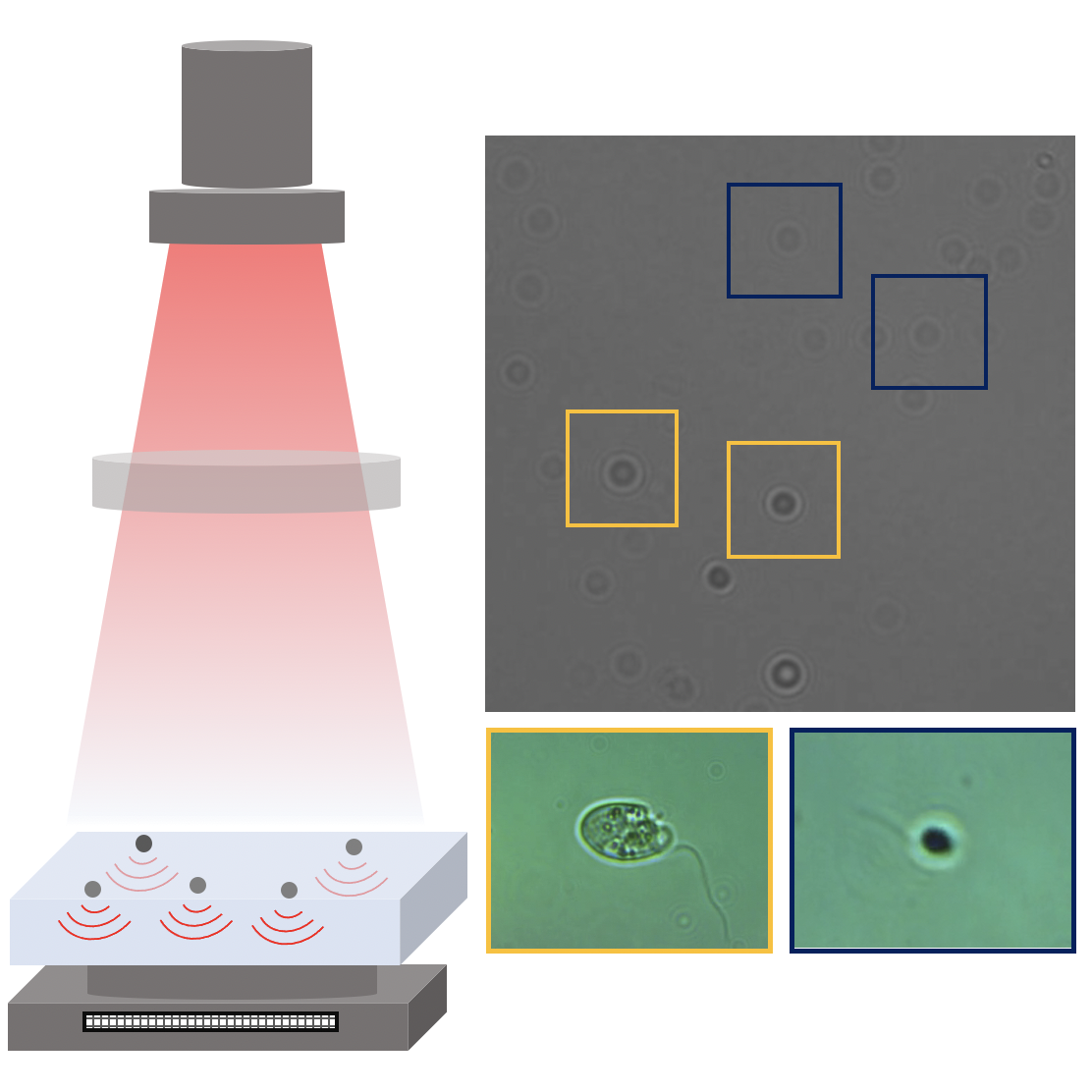
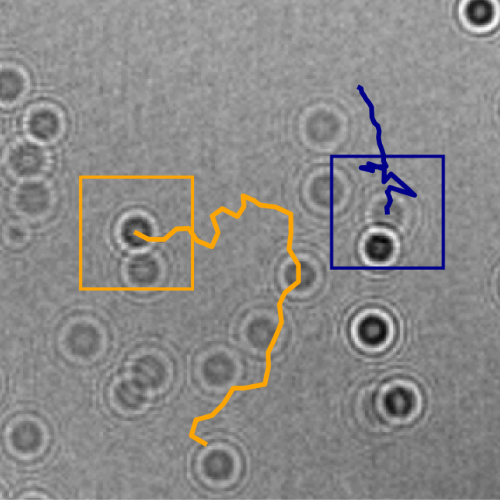
Here, we demonstrate that by combining holographic microscopy with deep learning, we can follow microplanktons through generations, by continuously measuring their three dimensional position and dry mass. The deep learning algorithms circumvent the computationally intensive processing of holographic data, and allow measurements over extended periods of time. This permits us to reliably estimate growth rates, both in terms of dry mass increase and cell divisions, as well as to measure trophic interactions between species such as predation events. We exemplify this by detailed descriptions of microzooplankton feeding events, cell divisions, and long term monitoring of single cells from division to division.