Measuring translational and rotational dynamics of colloid nanoparticles at the nanoscale with liquid-phase transmission electron microscopy
Murat Nulati Yesibolati, Assistant professor, Technical University of Denmark
4 August 2022, 10:30 CEST
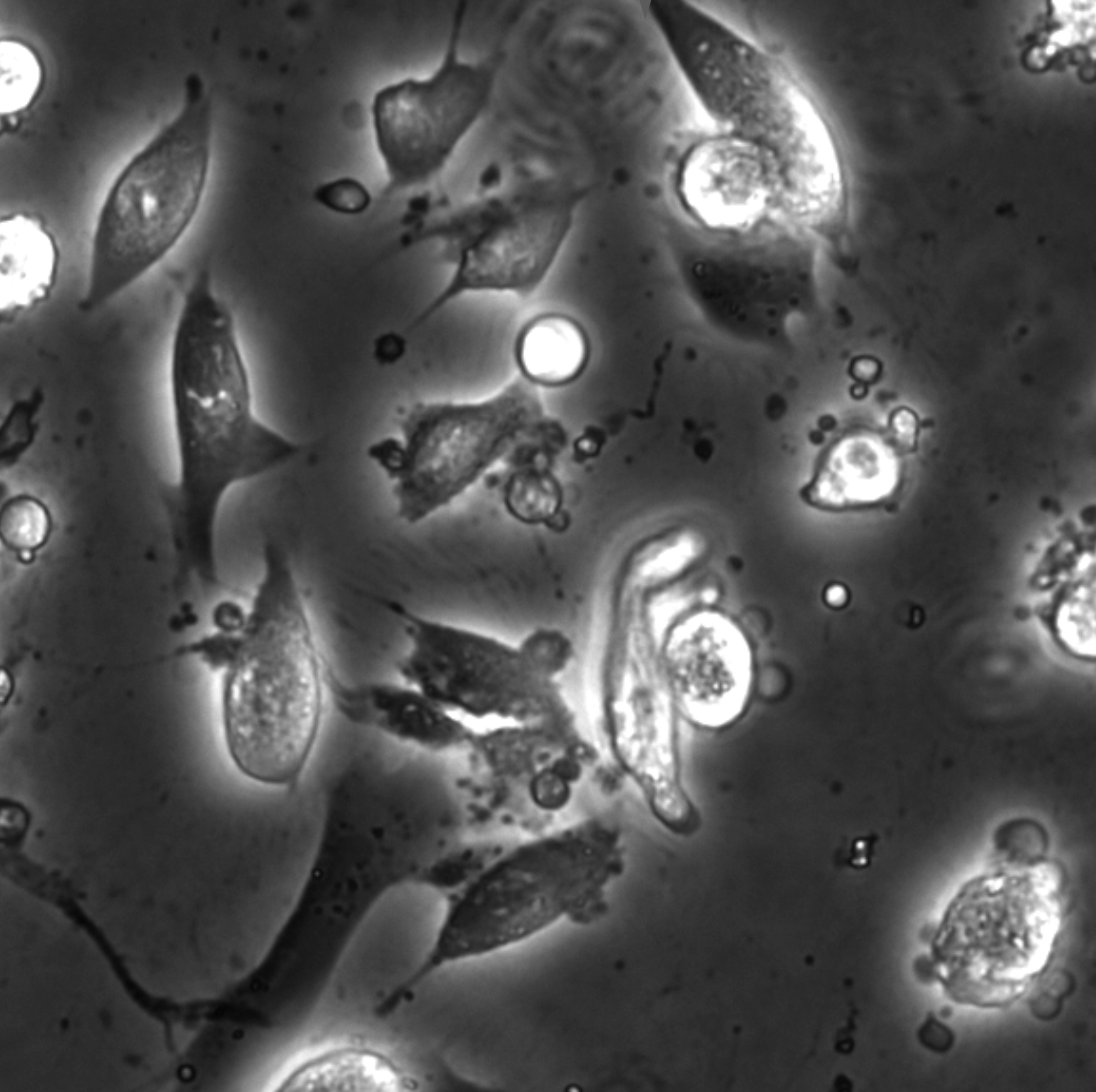
How nanoparticles (NPs) in a liquid suspension grow, transport, and interact with each other and surrounding interfaces are of fundamental interest in the colloidal matter, biomedical applications, microfluidics, and artificial micro/nanoscopic motors. Traditionally, imaging of such liquid processes has been limited to optical microscopy (OM). Bulk-level methods such as conventional OM and light scattering methods such as dynamic light scattering (DLS) cannot deliver nanometer spatial resolution at the single-particle level. Recently, liquid-phase transmission electron microscopy (LPTEM) [1] has revolutionized the access to the nanoscale, label-free imaging of a wide variety of liquid processes. Typically, the liquid cells used for LPTEM consist of electron-transparent silicon nitride (SiNx) windows suspended on two Si chips, which enclose a liquid sample layer with a thickness ranging from a few hundred nanometers to a couple of microns. With LPTEM, NP dynamics, such as nucleation and growth, self-assembly, and interactions, have been studied with sub-nanometer spatial resolution and millisecond temporal resolution.
We demonstrate how LPTEM can be used to measure the motion of individual NPs and agglomerates. Only at low electron flux do we find that individual NPs exhibit Brownian motion consistent with optical control experiments and theoretical predictions for unhindered passive diffusive motion in bulk liquids [2]. For increasing electron flux, we find increasingly faster than passive motion that still appears effectively Brownian. We discuss the possible origins of this beam–sample interaction. This establishes conditions for the use of LPTEM as a reliable tool for imaging nanoscale hydrodynamics at the nanoscale.
Bio
Murat N. Yesibolati is an Assistant Professor at Technical University of Denmark (DTU), Denmark. Murat defended his Ph.D. thesis titled “Electron holography and particle dynamics in liquid phase transmission electron microscopy” at DTU in 06.2018 under the supervision of Prof. Kristian Mølhave, DTU. Currently, he is focusing on developing a novel nanochannel liquid cell and exploring mass transport in nanochannels using advanced transmission electron microscopy. His research was supported by the Technical University of Denmark, by the Danish Research Council for Technology, grant no. 12-126194, the Advanced Materials for Energy-Water Systems (AMEWS) Center, Office of Science, Basic Energy Sciences, USA, grant number DE-AC02-06CH11357, and the VILLUM foundation, grant number 00028273.
References
[1] de Jonge, N. and F.M. Ross, Electron microscopy of specimens in liquid. Nature Nanotechnology, 2011. 6: p. 695.
[2] Yesibolati, M.N., et al., Unhindered Brownian Motion of Individual Nanoparticles in Liquid-Phase Scanning Transmission Electron Microscopy. Nano Letters, 2020. 20(10): p. 7108-7115.
Place: Nexus
Date: 4 August 2022
Time: 10:30 CEST