Jesús Pineda
Date: 23 August 2023
Time: 2:30 PM PDT
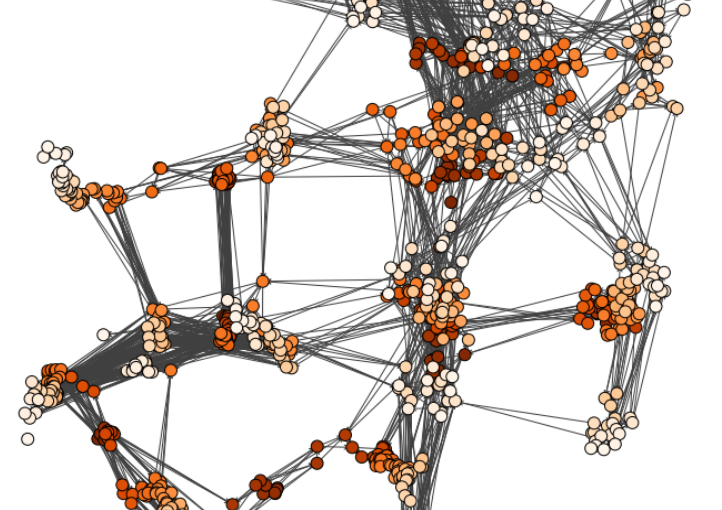
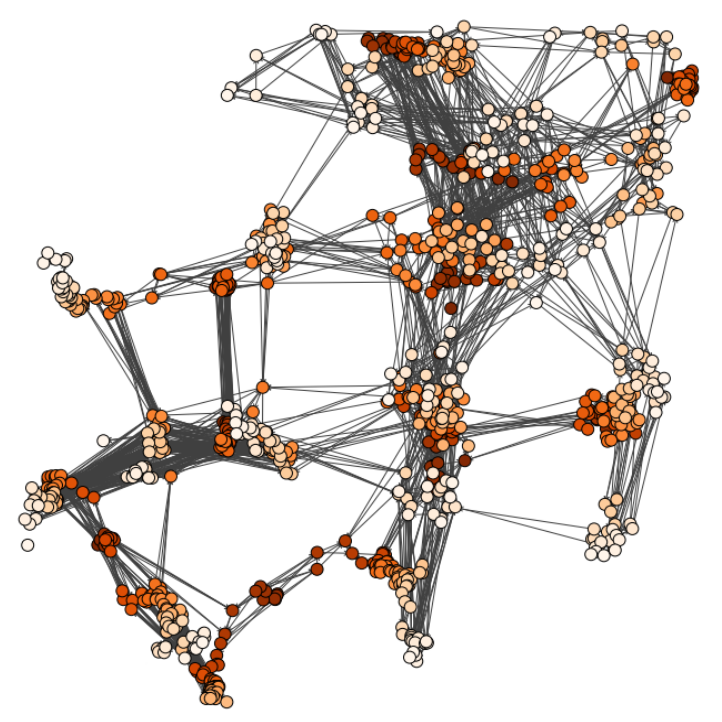
Characterizing dynamic processes in living systems provides essential information for advancing our understanding of life processes in health and diseases and for developing new technologies and treatments. In the past two decades, optical microscopy has undergone significant developments, enabling us to study the motion of cells, organelles, and individual molecules with unprecedented detail at various scales in space and time. However, analyzing the dynamic processes that occur in complex and crowded environments remains a challenge. This work introduces MAGIK, a deep-learning framework for the analysis of biological system dynamics from time-lapse microscopy. MAGIK models the movement and interactions of particles through a directed graph where nodes represent detections and edges connect spatiotemporally close nodes. The framework utilizes an attention-based graph neural network (GNN) to process the graph and modulate the strength of associations between its elements, enabling MAGIK to derive insights into the dynamics of the systems. MAGIK provides a key enabling technology to estimate any dynamic aspect of the particles, from reconstructing their trajectories to inferring local and global dynamics. We demonstrate the flexibility and reliability of the framework by applying it to real and simulated data corresponding to a broad range of biological experiments.
Reference
Pineda, J., Midtvedt, B., Bachimanchi, H. et al. Geometric deep learning reveals the spatiotemporal features of microscopic motion. Nat Mach Intell 5, 71–82 (2023)